Endogenous Viral Element-Derived Piwi-Interacting RNAs (piRNAs) Are Not Required for Production of Ping-Pong-Dependent piRNAs from Diaphorina citri Densovirus
- PMID: 32994324
- PMCID: PMC7527727
- DOI: 10.1128/mBio.02209-20
Endogenous Viral Element-Derived Piwi-Interacting RNAs (piRNAs) Are Not Required for Production of Ping-Pong-Dependent piRNAs from Diaphorina citri Densovirus
Abstract
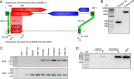
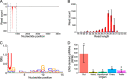
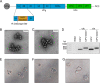
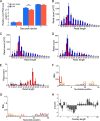
Piwi-interacting RNAs (piRNAs) are a class of small RNAs primarily responsible for silencing transposons in the animal germ line. The ping-pong cycle, the posttranscriptional silencing branch of the piRNA pathway, relies on piRNAs produced from endogenous transposon remnants to direct cleavage of transposon RNA via association with Piwi-family Argonaute proteins. In some mosquito species and mosquito-derived cell lines expressing a functionally expanded group of Piwi-family Argonaute proteins, both RNA and DNA viruses are targeted by piRNAs in a manner thought to involve direct processing of exogenous viral RNA into piRNAs. Whether viruses are targeted by piRNAs in nonmosquito species is unknown. Partial integrations of DNA and nonretroviral RNA virus genomes, termed endogenous viral elements (EVEs), are abundant in arthropod genomes and often produce piRNAs that are speculated to target cognate viruses through the ping-pong cycle. Here, we describe a Diaphorina citri densovirus (DcDV)-derived EVE in the genome of Diaphorina citri We found that this EVE gives rise to DcDV-specific primary piRNAs and is unevenly distributed among D. citri populations. Unexpectedly, we found that DcDV is targeted by ping-pong-dependent virus-derived piRNAs (vpiRNAs) in D. citri lacking the DcDV-derived EVE, while four naturally infecting RNA viruses of D. citri are not targeted by vpiRNAs. Furthermore, a recombinant Cricket paralysis virus containing a portion of the DcDV genome corresponding to the DcDV-derived EVE was not targeted by vpiRNAs during infection in D. citri harboring the EVE. These results demonstrate that viruses can be targeted by piRNAs in a nonmosquito species independently of endogenous piRNAs.IMPORTANCE Small RNAs serve as specificity determinants of antiviral responses in insects. Piwi-interacting RNAs (piRNAs) are a class of small RNAs found in animals, and their primary role is to direct antitransposon responses. These responses require endogenous piRNAs complementary to transposon RNA. Additionally, piRNAs have been shown to target RNA and DNA viruses in some mosquito species. In contrast to transposons, targeting of viruses by the piRNA pathway in these mosquito species does not require endogenous piRNAs. Here, we show that piRNAs target a DNA virus, but not RNA viruses, in an agricultural insect pest. We found that targeting of this DNA virus did not require endogenous piRNAs and that endogenous piRNAs did not mediate targeting of an RNA virus with which they shared complementary sequence. Our results highlight differences between mosquitoes and our experimental system and raise the possibility that DNA viruses may be targeted by piRNAs in other species.
Keywords: Diaphorina citri; RNA interference; densovirus; piRNA; small RNA.
Copyright © 2020 Nigg et al.
Figures

Similar articles
-
Endogenous Viral Elements Are Widespread in Arthropod Genomes and Commonly Give Rise to PIWI-Interacting RNAs.J Virol. 2019 Mar 5;93(6):e02124-18. doi: 10.1128/JVI.02124-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30567990 Free PMC article.
-
Deducing the Role of Virus Genome-Derived PIWI-Associated RNAs in the Mosquito-Arbovirus Arms Race.Front Genet. 2019 Nov 28;10:1114. doi: 10.3389/fgene.2019.01114. eCollection 2019. Front Genet. 2019. PMID: 31850054 Free PMC article. Review.
-
Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells.Nucleic Acids Res. 2015 Jul 27;43(13):6545-56. doi: 10.1093/nar/gkv590. Epub 2015 Jun 11. Nucleic Acids Res. 2015. PMID: 26068474 Free PMC article.
-
Diaphorina citri densovirus is a persistently infecting virus with a hybrid genome organization and unique transcription strategy.J Gen Virol. 2020 Feb;101(2):226-239. doi: 10.1099/jgv.0.001371. Epub 2019 Dec 19. J Gen Virol. 2020. PMID: 31855134
-
Virus and endogenous viral element-derived small non-coding RNAs and their roles in insect-virus interaction.Curr Opin Insect Sci. 2022 Feb;49:85-92. doi: 10.1016/j.cois.2021.12.007. Epub 2021 Dec 30. Curr Opin Insect Sci. 2022. PMID: 34974161 Review.
Cited by
-
Untangling an insect's virome from its endogenous viral elements.BMC Genomics. 2023 Oct 24;24(1):636. doi: 10.1186/s12864-023-09737-z. BMC Genomics. 2023. PMID: 37875824 Free PMC article.
-
What Are the Functional Roles of Piwi Proteins and piRNAs in Insects?Insects. 2023 Feb 14;14(2):187. doi: 10.3390/insects14020187. Insects. 2023. PMID: 36835756 Free PMC article. Review.
-
High-quality, chromosome-scale genome assemblies: comparisons of three Diaphorina citri (Asian citrus psyllid) geographic populations.DNA Res. 2022 Jun 25;29(4):dsac027. doi: 10.1093/dnares/dsac027. DNA Res. 2022. PMID: 35866687 Free PMC article.
-
Transcriptome Analysis Reveals a Diverse Range of Novel Viruses in Australian Sugarcane Soldier Fly (Inopus flavus) Larvae.Viruses. 2024 Mar 27;16(4):516. doi: 10.3390/v16040516. Viruses. 2024. PMID: 38675859 Free PMC article.
-
Non-gonadal somatic piRNA pathways ensure sexual differentiation, larval growth, and wing development in silkworms.PLoS Genet. 2023 Sep 21;19(9):e1010912. doi: 10.1371/journal.pgen.1010912. eCollection 2023 Sep. PLoS Genet. 2023. PMID: 37733654 Free PMC article.
References
-
- Dietrich I, Jansen S, Fall G, Lorenzen S, Rudolf M, Huber K, Heitmann A, Schicht S, Ndiaye EH, Watson M, Castelli I, Brennan B, Elliott RM, Diallo M, Sall AA, Failloux A-B, Schnettler E, Kohl A, Becker SC. 2017. RNA interference restricts Rift Valley Fever virus in multiple insect systems. MSphere 2:e00090-17. doi:10.1128/mSphere.00090-17. - DOI - PMC - PubMed
-
- Varjak M, Maringer K, Watson M, Sreenu VB, Fredericks AC, Pondeville E, Donald CL, Sterk J, Kean J, Vazeille M, Failloux A-B, Kohl A, Schnettler E. 2017. Aedes aegypti Piwi4 is a noncanonical PIWI protein involved in antiviral responses. MSphere 2:e00144-17. doi:10.1128/mSphere.00144-17. - DOI - PMC - PubMed

