Engineered TALE Repeats for Enhanced Imaging-Based Analysis of Cellular 5-Methylcytosine
- PMID: 32991020
- PMCID: PMC7894354
- DOI: 10.1002/cbic.202000563
Engineered TALE Repeats for Enhanced Imaging-Based Analysis of Cellular 5-Methylcytosine
Abstract
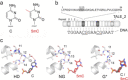
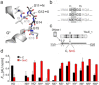
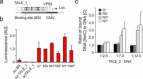
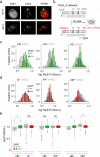
Transcription-activator-like effectors (TALEs) are repeat-based, programmable DNA-binding proteins that can be engineered to recognize sequences of canonical and epigenetically modified nucleobases. Fluorescent TALEs can be used for the imaging-based analysis of cellular 5-methylcytosine (5 mC) in repetitive DNA sequences. This is based on recording fluorescence ratios from cell co-stains with two TALEs: an analytical TALE targeting the cytosine (C) position of interest through a C-selective repeat that is blocked by 5 mC, and a control TALE targeting the position with a universal repeat that binds both C and 5 mC. To enhance this approach, we report herein the development of novel 5 mC-selective repeats and their integration into TALEs that can replace universal TALEs in imaging-based 5 mC analysis, resulting in a methylation-dependent response of both TALEs. We screened a library of size-reduced repeats and identified several 5 mC binders. Compared to the 5 mC-binding repeat of natural TALEs and to the universal repeat, two repeats containing aromatic residues showed enhancement of 5 mC binding and selectivity in cellular transcription activation and electromobility shift assays, respectively. In co-stains of cellular SATIII DNA with a corresponding C-selective TALE, this selectivity results in a positive methylation response of the new TALE, offering perspectives for studying 5 mC functions in chromatin regulation by in situ imaging with increased dynamic range.
Keywords: DNA methylation; epigenetics; imaging probes; membrane-less organelles.
© 2020 The Authors. ChemBioChem published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Achieving single-nucleotide resolution of 5-methylcytosine detection with TALEs.Chembiochem. 2015 Jan 19;16(2):228-31. doi: 10.1002/cbic.201402408. Epub 2014 Dec 17. Chembiochem. 2015. PMID: 25522353
-
Selective recognition of N4-methylcytosine in DNA by engineered transcription-activator-like effectors.Philos Trans R Soc Lond B Biol Sci. 2018 Jun 5;373(1748):20170078. doi: 10.1098/rstb.2017.0078. Philos Trans R Soc Lond B Biol Sci. 2018. PMID: 29685980 Free PMC article.
-
Complete, Programmable Decoding of Oxidized 5-Methylcytosine Nucleobases in DNA by Chemoselective Blockage of Universal Transcription-Activator-Like Effector Repeats.J Am Chem Soc. 2018 May 9;140(18):5904-5908. doi: 10.1021/jacs.8b02909. Epub 2018 Apr 26. J Am Chem Soc. 2018. PMID: 29677450
-
TALEored Epigenetics: A DNA-Binding Scaffold for Programmable Epigenome Editing and Analysis.Chembiochem. 2016 Jun 2;17(11):975-80. doi: 10.1002/cbic.201600072. Epub 2016 Apr 20. Chembiochem. 2016. PMID: 26972580 Review.
-
Modified nucleobase-specific gene regulation using engineered transcription activator-like effectors.Adv Drug Deliv Rev. 2019 Jul;147:59-65. doi: 10.1016/j.addr.2019.08.011. Epub 2019 Sep 9. Adv Drug Deliv Rev. 2019. PMID: 31513826 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

