TDP-43 dysfunction results in R-loop accumulation and DNA replication defects
- PMID: 32989039
- PMCID: PMC7648616
- DOI: 10.1242/jcs.244129
TDP-43 dysfunction results in R-loop accumulation and DNA replication defects
Abstract
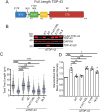
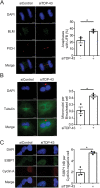
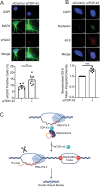
TAR DNA-binding protein 43 (TDP-43; also known as TARDBP) is an RNA-binding protein whose aggregation is a hallmark of the neurodegenerative disorders amyotrophic lateral sclerosis and frontotemporal dementia. TDP-43 loss increases DNA damage and compromises cell viability, but the actual function of TDP-43 in preventing genome instability remains unclear. Here, we show that loss of TDP-43 increases R-loop formation in a transcription-dependent manner and results in DNA replication stress. TDP-43 nucleic-acid-binding and self-assembly activities are important in inhibiting R-loop accumulation and preserving normal DNA replication. We also found that TDP-43 cytoplasmic aggregation impairs TDP-43 function in R-loop regulation. Furthermore, increased R-loop accumulation and DNA damage is observed in neurons upon loss of TDP-43. Together, our findings indicate that TDP-43 function and normal protein homeostasis are crucial in maintaining genomic stability through a co-transcriptional process that prevents aberrant R-loop accumulation. We propose that the increased R-loop formation and genomic instability associated with TDP-43 loss are linked to the pathogenesis of TDP-43 proteinopathies.This article has an associated First Person interview with the first author of the paper.
Keywords: DNA Replication; R-loops; RNA:DNA hybrids; TARDBP; TDP-43.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures
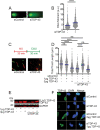
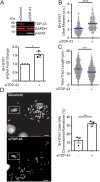
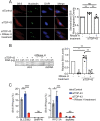
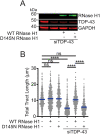

Similar articles
-
TDP-43 mutations link Amyotrophic Lateral Sclerosis with R-loop homeostasis and R loop-mediated DNA damage.PLoS Genet. 2020 Dec 10;16(12):e1009260. doi: 10.1371/journal.pgen.1009260. eCollection 2020 Dec. PLoS Genet. 2020. PMID: 33301444 Free PMC article.
-
Quantification of the Relative Contributions of Loss-of-function and Gain-of-function Mechanisms in TAR DNA-binding Protein 43 (TDP-43) Proteinopathies.J Biol Chem. 2016 Sep 9;291(37):19437-48. doi: 10.1074/jbc.M116.737726. Epub 2016 Jul 21. J Biol Chem. 2016. PMID: 27445339 Free PMC article.
-
Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins.J Neurochem. 2016 Aug;138 Suppl 1:95-111. doi: 10.1111/jnc.13625. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27015757 Review.
-
TDP-43 proteinopathies: pathological identification of brain regions differentiating clinical phenotypes.Brain. 2015 Oct;138(Pt 10):3110-22. doi: 10.1093/brain/awv220. Epub 2015 Jul 31. Brain. 2015. PMID: 26231953
-
TDP-43 pathology: From noxious assembly to therapeutic removal.Prog Neurobiol. 2022 Apr;211:102229. doi: 10.1016/j.pneurobio.2022.102229. Epub 2022 Jan 29. Prog Neurobiol. 2022. PMID: 35101542 Review.
Cited by
-
Ataxin-2 gene: a powerful modulator of neurological disorders.Curr Opin Neurol. 2021 Aug 1;34(4):578-588. doi: 10.1097/WCO.0000000000000959. Curr Opin Neurol. 2021. PMID: 34010218 Free PMC article. Review.
-
The chromatin-associated lncREST ensures effective replication stress response by promoting the assembly of fork signaling factors.Nat Commun. 2024 Feb 1;15(1):978. doi: 10.1038/s41467-024-45183-5. Nat Commun. 2024. PMID: 38302450 Free PMC article.
-
Walking a tightrope: The complex balancing act of R-loops in genome stability.Mol Cell. 2022 Jun 16;82(12):2267-2297. doi: 10.1016/j.molcel.2022.04.014. Epub 2022 May 3. Mol Cell. 2022. PMID: 35508167 Free PMC article. Review.
-
The chromatin network helps prevent cancer-associated mutagenesis at transcription-replication conflicts.Nat Commun. 2023 Oct 28;14(1):6890. doi: 10.1038/s41467-023-42653-0. Nat Commun. 2023. PMID: 37898641 Free PMC article.
-
New Faces of old Friends: Emerging new Roles of RNA-Binding Proteins in the DNA Double-Strand Break Response.Front Mol Biosci. 2021 May 7;8:668821. doi: 10.3389/fmolb.2021.668821. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026839 Free PMC article. Review.
References
-
- Afroz T., Hock E.-M., Ernst P., Foglieni C., Jambeau M., Gilhespy L. A. B., Laferriere F., Maniecka Z., Plückthun A., Mittl P. et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat. Commun. 8, 45 10.1038/s41467-017-00062-0 - DOI - PMC - PubMed
-
- Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y. et al. (2006). TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602-611. 10.1016/j.bbrc.2006.10.093 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

