Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions
- PMID: 32988401
- PMCID: PMC7523332
- DOI: 10.1186/s13073-020-00776-9
Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions
Abstract
Background: Solid tumors such as pancreatic ductal adenocarcinoma (PDAC) comprise not just tumor cells but also a microenvironment with which the tumor cells constantly interact. Detailed characterization of the cellular composition of the tumor microenvironment is critical to the understanding of the disease and treatment of the patient. Single-cell transcriptomics has been used to study the cellular composition of different solid tumor types including PDAC. However, almost all of those studies used primary tumor tissues.
Methods: In this study, we employed a single-cell RNA sequencing technology to profile the transcriptomes of individual cells from dissociated primary tumors or metastatic biopsies obtained from patients with PDAC. Unsupervised clustering analysis as well as a new supervised classification algorithm, SuperCT, was used to identify the different cell types within the tumor tissues. The expression signatures of the different cell types were then compared between primary tumors and metastatic biopsies. The expressions of the cell type-specific signature genes were also correlated with patient survival using public datasets.
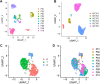
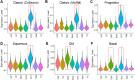
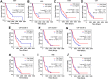
Results: Our single-cell RNA sequencing analysis revealed distinct cell types in primary and metastatic PDAC tissues including tumor cells, endothelial cells, cancer-associated fibroblasts (CAFs), and immune cells. The cancer cells showed high inter-patient heterogeneity, whereas the stromal cells were more homogenous across patients. Immune infiltration varies significantly from patient to patient with majority of the immune cells being macrophages and exhausted lymphocytes. We found that the tumor cellular composition was an important factor in defining the PDAC subtypes. Furthermore, the expression levels of cell type-specific markers for EMT+ cancer cells, activated CAFs, and endothelial cells significantly associated with patient survival.
Conclusions: Taken together, our work identifies significant heterogeneity in cellular compositions of PDAC tumors and between primary tumors and metastatic lesions. Furthermore, the cellular composition was an important factor in defining PDAC subtypes and significantly correlated with patient outcome. These findings provide valuable insights on the PDAC microenvironment and could potentially inform the management of PDAC patients.
Keywords: Cellular heterogeneity; Pancreatic cancer; Pancreatic cancer subtypes; Single-cell RNA sequencing.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Inter- and intra-tumoural heterogeneity in cancer-associated fibroblasts of human pancreatic ductal adenocarcinoma.J Pathol. 2019 May;248(1):51-65. doi: 10.1002/path.5224. Epub 2019 Feb 22. J Pathol. 2019. PMID: 30575030 Free PMC article.
-
Deciphering the Prognostic Implications of the Components and Signatures in the Immune Microenvironment of Pancreatic Ductal Adenocarcinoma.Front Immunol. 2021 Mar 10;12:648917. doi: 10.3389/fimmu.2021.648917. eCollection 2021. Front Immunol. 2021. PMID: 33777046 Free PMC article.
-
Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression.EBioMedicine. 2021 Apr;66:103315. doi: 10.1016/j.ebiom.2021.103315. Epub 2021 Apr 2. EBioMedicine. 2021. PMID: 33819739 Free PMC article.
-
[Metastasis of pancreatic tumors].Pathologe. 2015 Nov;36 Suppl 2:176-80. doi: 10.1007/s00292-015-0077-0. Pathologe. 2015. PMID: 26391249 Review. German.
-
CAFs-Associated Genes (CAFGs) in Pancreatic Ductal Adenocarcinoma (PDAC) and Novel Therapeutic Strategy.Int J Mol Sci. 2024 May 30;25(11):6003. doi: 10.3390/ijms25116003. Int J Mol Sci. 2024. PMID: 38892190 Free PMC article. Review.
Cited by
-
Necroptosis activation is associated with greater methylene blue-photodynamic therapy-induced cytotoxicity in human pancreatic ductal adenocarcinoma cells.Photochem Photobiol Sci. 2023 Apr;22(4):729-744. doi: 10.1007/s43630-022-00347-4. Epub 2022 Dec 10. Photochem Photobiol Sci. 2023. PMID: 36495407
-
Inhibition of insulin-like growth factors increases production of CXCL9/10 by macrophages and fibroblasts and facilitates CD8+ cytotoxic T cell recruitment to pancreatic tumours.Front Immunol. 2024 Aug 5;15:1382538. doi: 10.3389/fimmu.2024.1382538. eCollection 2024. Front Immunol. 2024. PMID: 39165364 Free PMC article.
-
Construction of a radiogenomic association map of pancreatic ductal adenocarcinoma.BMC Cancer. 2023 Feb 27;23(1):189. doi: 10.1186/s12885-023-10658-z. BMC Cancer. 2023. PMID: 36843111 Free PMC article.
-
scDrug: From single-cell RNA-seq to drug response prediction.Comput Struct Biotechnol J. 2022 Dec 1;21:150-157. doi: 10.1016/j.csbj.2022.11.055. eCollection 2023. Comput Struct Biotechnol J. 2022. PMID: 36544472 Free PMC article.
-
Single-cell RNA-seq analysis reveals BHLHE40-driven pro-tumour neutrophils with hyperactivated glycolysis in pancreatic tumour microenvironment.Gut. 2023 May;72(5):958-971. doi: 10.1136/gutjnl-2021-326070. Epub 2022 Jun 10. Gut. 2023. PMID: 35688610 Free PMC article.
References
-
- Hingorani SR, Zheng L, Bullock AJ, Seery TE, Harris WP, Sigal DS, et al. HALO 202: randomized phase II study of PEGPH20 plus nab-paclitaxel/gemcitabine versus nab-paclitaxel/gemcitabine in patients with untreated, metastatic pancreatic ductal adenocarcinoma. J Clin Oncol. 2018;36(4):359–366. doi: 10.1200/JCO.2017.74.9564. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

