Positive Role of the MHC Class-I Antigen Presentation Regulator m04/gp34 of Murine Cytomegalovirus in Antiviral Protection by CD8 T Cells
- PMID: 32984075
- PMCID: PMC7479846
- DOI: 10.3389/fcimb.2020.00454
Positive Role of the MHC Class-I Antigen Presentation Regulator m04/gp34 of Murine Cytomegalovirus in Antiviral Protection by CD8 T Cells
Abstract
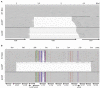
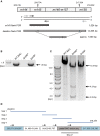
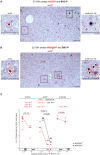
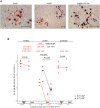
Murine cytomegalovirus (mCMV) codes for MHC class-I trafficking modulators m04/gp34, m06/gp48, and m152/gp40. By interacting with the MHC class-Iα chain, these proteins disconnect peptide-loaded MHC class-I (pMHC-I) complexes from the constitutive vesicular flow to the cell surface. Based on the assumption that all three inhibit antigen presentation, and thus the recognition of infected cells by CD8 T cells, they were referred to as "immunoevasins." Improved antigen presentation mediated by m04 in the presence of m152 after infection with deletion mutant mCMV-Δm06W, compared to mCMV-Δm04m06 expressing only m152, led us to propose renaming these molecules "viral regulators of antigen presentation" (vRAP) to account for both negative and positive functions. In accordance with a positive function, m04-pMHC-I complexes were found to be displayed on the cell surface, where they are primarily known as ligands for Ly49 family natural killer (NK) cell receptors. Besides the established role of m04 in NK cell silencing or activation, an anti-immunoevasive function by activation of CD8 T cells is conceivable, because the binding site of m04 to MHC class-Iα appears not to mask the peptide binding site for T-cell receptor recognition. However, functional evidence was based on mCMV-Δm06W, a virus of recently doubted authenticity. Here we show that mCMV-Δm06W actually represents a mixture of an authentic m06 deletion mutant and a mutant with an accidental additional deletion of a genome region encompassing also gene m152. Reanalysis of previously published experiments for the authentic mutant in the mixture confirms the previously concluded positive vRAP function of m04.
Keywords: BAC mutagenesis; CD8 T cells; adoptive cell transfer; antigen presentation; immune evasion; immunoevasin; next-generation sequencing (NGS); recombinant virus.
Copyright © 2020 Becker, Fink, Podlech, Giese, Schmiedeke, Bukur, Reddehase and Lemmermann.
Figures
Similar articles
-
Cytomegalovirus immune evasion sets the functional avidity threshold for protection by CD8 T cells.Med Microbiol Immunol. 2023 Apr;212(2):153-163. doi: 10.1007/s00430-022-00733-w. Epub 2022 Apr 1. Med Microbiol Immunol. 2023. PMID: 35364731 Free PMC article. Review.
-
The putative natural killer decoy early gene m04 (gp34) of murine cytomegalovirus encodes an antigenic peptide recognized by protective antiviral CD8 T cells.J Virol. 2000 Feb;74(4):1871-84. doi: 10.1128/jvi.74.4.1871-1884.2000. J Virol. 2000. PMID: 10644360 Free PMC article.
-
Cytomegalovirus encodes a positive regulator of antigen presentation.J Virol. 2006 Aug;80(15):7613-24. doi: 10.1128/JVI.00723-06. J Virol. 2006. PMID: 16840340 Free PMC article.
-
The role of NKG2D signaling in inhibition of cytotoxic T-lymphocyte lysis by the Murine cytomegalovirus immunoevasin m152/gp40.J Virol. 2007 Nov;81(22):12564-71. doi: 10.1128/JVI.01328-07. Epub 2007 Sep 12. J Virol. 2007. PMID: 17855532 Free PMC article.
-
Cytomegaloviral control of MHC class I function in the mouse.Immunol Rev. 1999 Apr;168:167-76. doi: 10.1111/j.1600-065x.1999.tb01291.x. Immunol Rev. 1999. PMID: 10399073 Review.
Cited by
-
Oncolytic α-herpesvirus and myeloid-tropic cytomegalovirus cooperatively enhance systemic antitumor responses.Mol Ther. 2024 Jan 3;32(1):241-256. doi: 10.1016/j.ymthe.2023.11.003. Epub 2023 Nov 4. Mol Ther. 2024. PMID: 37927036 Free PMC article.
-
Therapeutic Vaccination of Hematopoietic Cell Transplantation Recipients Improves Protective CD8 T-Cell Immunotherapy of Cytomegalovirus Infection.Front Immunol. 2021 Aug 19;12:694588. doi: 10.3389/fimmu.2021.694588. eCollection 2021. Front Immunol. 2021. PMID: 34489940 Free PMC article.
-
Cytomegalovirus immune evasion sets the functional avidity threshold for protection by CD8 T cells.Med Microbiol Immunol. 2023 Apr;212(2):153-163. doi: 10.1007/s00430-022-00733-w. Epub 2022 Apr 1. Med Microbiol Immunol. 2023. PMID: 35364731 Free PMC article. Review.
-
Stochastic Episodes of Latent Cytomegalovirus Transcription Drive CD8 T-Cell "Memory Inflation" and Avoid Immune Evasion.Front Immunol. 2021 Apr 22;12:668885. doi: 10.3389/fimmu.2021.668885. eCollection 2021. Front Immunol. 2021. PMID: 33968074 Free PMC article.
-
Direct antigen presentation is the canonical pathway of cytomegalovirus CD8 T-cell priming regulated by balanced immune evasion ensuring a strong antiviral response.Front Immunol. 2023 Dec 12;14:1272166. doi: 10.3389/fimmu.2023.1272166. eCollection 2023. Front Immunol. 2023. PMID: 38149242 Free PMC article.
References
-
- Babic M., Pyzik M., Zafirova B., Mitrovic M., Butorac V., Lanier L. L., et al. . (2010). Cytomegalovirus immunoevasin reveals the physiological role of “missing self” recognition in natural killer cell dependent virus control in vivo. J. Exp. Med. 207, 2663–2673. 10.1084/jem.20100921 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

