A blood-based host gene expression assay for early detection of respiratory viral infection: an index-cluster prospective cohort study
- PMID: 32979932
- PMCID: PMC7515566
- DOI: 10.1016/S1473-3099(20)30486-2
A blood-based host gene expression assay for early detection of respiratory viral infection: an index-cluster prospective cohort study
Abstract
Background: Early and accurate identification of individuals with viral infections is crucial for clinical management and public health interventions. We aimed to assess the ability of transcriptomic biomarkers to identify naturally acquired respiratory viral infection before typical symptoms are present.
Methods: In this index-cluster study, we prospectively recruited a cohort of undergraduate students (aged 18-25 years) at Duke University (Durham, NC, USA) over a period of 5 academic years. To identify index cases, we monitored students for the entire academic year, for the presence and severity of eight symptoms of respiratory tract infection using a daily web-based survey, with symptoms rated on a scale of 0-4. Index cases were defined as individuals who reported a 6-point increase in cumulative daily symptom score. Suspected index cases were visited by study staff to confirm the presence of reported symptoms of illness and to collect biospecimen samples. We then identified clusters of close contacts of index cases (ie, individuals who lived in close proximity to index cases, close friends, and partners) who were presumed to be at increased risk of developing symptomatic respiratory tract infection while under observation. We monitored each close contact for 5 days for symptoms and viral shedding and measured transcriptomic responses at each timepoint each day using a blood-based 36-gene RT-PCR assay.
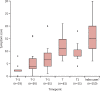
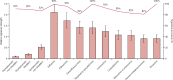
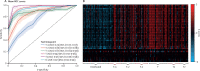
Findings: Between Sept 1, 2009, and April 10, 2015, we enrolled 1465 participants. Of 264 index cases with respiratory tract infection symptoms, 150 (57%) had a viral cause confirmed by RT-PCR. Of their 555 close contacts, 106 (19%) developed symptomatic respiratory tract infection with a proven viral cause during the observation window, of whom 60 (57%) had the same virus as their associated index case. Nine viruses were detected in total. The transcriptomic assay accurately predicted viral infection at the time of maximum symptom severity (mean area under the receiver operating characteristic curve [AUROC] 0·94 [95% CI 0·92-0·96]), as well as at 1 day (0·87 [95% CI 0·84-0·90]), 2 days (0·85 [0·82-0·88]), and 3 days (0·74 [0·71-0·77]) before peak illness, when symptoms were minimal or absent and 22 (62%) of 35 individuals, 25 (69%) of 36 individuals, and 24 (82%) of 29 individuals, respectively, had no detectable viral shedding.
Interpretation: Transcriptional biomarkers accurately predict and diagnose infection across diverse viral causes and stages of disease and thus might prove useful for guiding the administration of early effective therapy, quarantine decisions, and other clinical and public health interventions in the setting of endemic and pandemic infectious diseases.
Funding: US Defense Advanced Research Projects Agency.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Figures
Comment in
-
Recognising the asymptomatic enemy.Lancet Infect Dis. 2021 Mar;21(3):305-306. doi: 10.1016/S1473-3099(20)30587-9. Epub 2020 Sep 24. Lancet Infect Dis. 2021. PMID: 32979931 Free PMC article. No abstract available.
Similar articles
-
Onset and window of SARS-CoV-2 infectiousness and temporal correlation with symptom onset: a prospective, longitudinal, community cohort study.Lancet Respir Med. 2022 Nov;10(11):1061-1073. doi: 10.1016/S2213-2600(22)00226-0. Epub 2022 Aug 18. Lancet Respir Med. 2022. PMID: 35988572 Free PMC article.
-
Frequent detection of respiratory viruses in adult recipients of stem cell transplants with the use of real-time polymerase chain reaction, compared with viral culture.Clin Infect Dis. 2005 Mar 1;40(5):662-9. doi: 10.1086/427801. Epub 2005 Feb 7. Clin Infect Dis. 2005. PMID: 15714410 Free PMC article.
-
Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of reverse transcription-PCR DNA microarray systems.J Clin Microbiol. 2010 Nov;48(11):3836-42. doi: 10.1128/JCM.00733-10. Epub 2010 Aug 25. J Clin Microbiol. 2010. PMID: 20739481 Free PMC article.
-
Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis.Clin Microbiol Infect. 2018 Oct;24(10):1055-1063. doi: 10.1016/j.cmi.2017.11.018. Epub 2017 Dec 5. Clin Microbiol Infect. 2018. PMID: 29208560 Free PMC article. Review.
-
Community respiratory viruses in individuals with human immunodeficiency virus infection.Am J Med. 1997 Mar 17;102(3A):19-24; discussion 25-6. doi: 10.1016/s0002-9343(97)80005-8. Am J Med. 1997. PMID: 10868138 Free PMC article. Review.
Cited by
-
Dysregulated transcriptional responses to SARS-CoV-2 in the periphery support novel diagnostic approaches.medRxiv [Preprint]. 2020 Jul 26:2020.07.20.20155507. doi: 10.1101/2020.07.20.20155507. medRxiv. 2020. Update in: Nat Commun. 2021 Feb 17;12(1):1079. doi: 10.1038/s41467-021-21289-y PMID: 32743603 Free PMC article. Updated. Preprint.
-
A T-Cell-Derived 3-Gene Signature Distinguishes SARS-CoV-2 from Common Respiratory Viruses.Viruses. 2024 Jun 26;16(7):1029. doi: 10.3390/v16071029. Viruses. 2024. PMID: 39066192 Free PMC article.
-
TIPICO XI: report of the first series and podcast on infectious diseases and vaccines (aTIPICO).Hum Vaccin Immunother. 2021 Nov 2;17(11):4299-4327. doi: 10.1080/21645515.2021.1953351. Epub 2021 Nov 11. Hum Vaccin Immunother. 2021. PMID: 34762551 Free PMC article.
-
Selection of Ideal Reference Genes for Gene Expression Analysis in COVID-19 and Mucormycosis.Microbiol Spectr. 2022 Dec 21;10(6):e0165622. doi: 10.1128/spectrum.01656-22. Epub 2022 Nov 15. Microbiol Spectr. 2022. PMID: 36377893 Free PMC article.
-
Metavirome Analysis of Culex tritaeniorhynchus Reveals Novel Japanese Encephalitis Virus and Chikungunya Virus.Front Cell Infect Microbiol. 2022 Jun 30;12:938576. doi: 10.3389/fcimb.2022.938576. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35846772 Free PMC article.
References
-
- Shrestha SS, Swerdlow DL, Borse RH, et al. Estimating the burden of 2009 pandemic influenza A (H1N1) in the United States (April 2009–April 2010) Clin Infect Dis. 2011;52(suppl 1):S75–S82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

