Exploring Specific miRNA-mRNA Axes With Relationship to Taxanes-Resistance in Breast Cancer
- PMID: 32974144
- PMCID: PMC7473300
- DOI: 10.3389/fonc.2020.01397
Exploring Specific miRNA-mRNA Axes With Relationship to Taxanes-Resistance in Breast Cancer
Abstract
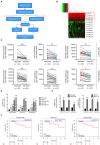
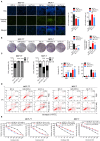
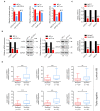
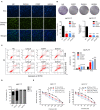
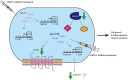
Breast cancer is the most prevalent type of malignancy in women worldwide. Taxanes (paclitaxel and docetaxel) are widely applied as first-line chemotherapeutic agents, while the therapeutic effect is seriously limited by the development of drug resistance. In the present study, we screened out several miRNAs dysregulated in taxanes-resistant breast cancer samples and confirmed that two miRNAs (miR-335-5p and let-7c-5p) played a major role in cell proliferation, apoptosis, and chemo-resistance. In addition, the weighted gene co-expression network analysis (WGCNA) for potential target genes of miR-335-5p and let-7c-5p identified three hub genes (CXCL9, CCR7, and SOCS1) with a positive relationship to taxanes-sensitivity. Further, target relationships between miR-335-5p and CXCL9, let-7c-5p and CCR7/SOCS1 were confirmed by dual-luciferase reporter assays. Importantly, the regulatory functions of CXCL9, CCR7, and SOCS1 on proliferation and chemoresistance were validated. In conclusion, our study shed light on clinical theragnostic relationships between miR-335-5p/CXCL9, let-7c-5p/CCR7/SOCS1 axes, and taxanes-resistance in breast cancer.
Keywords: WCGNA; breast cancer; let-7c-5p/CCR7/SOCS1 axis; miR-335-5p/CXCL9 axis; taxanes-resistance.
Copyright © 2020 Chen, Bao, Zhao, Yu, Zhong, Xu and Yan.
Figures


Similar articles
-
Cytological effects of honokiol treatment and its potential mechanism of action in non-small cell lung cancer.Biomed Pharmacother. 2019 Sep;117:109058. doi: 10.1016/j.biopha.2019.109058. Epub 2019 Jun 5. Biomed Pharmacother. 2019. PMID: 31176168
-
MicroRNAs as possible indicators of drug sensitivity in breast cancer cell lines.PLoS One. 2019 May 7;14(5):e0216400. doi: 10.1371/journal.pone.0216400. eCollection 2019. PLoS One. 2019. PMID: 31063487 Free PMC article.
-
Withaferin A mediated changes of miRNA expression in breast cancer-derived mammospheres.Mol Carcinog. 2022 Sep;61(9):876-889. doi: 10.1002/mc.23440. Epub 2022 Jun 30. Mol Carcinog. 2022. PMID: 35770722
-
miRNA Expression Profiling in Human Breast Cancer Diagnostics and Therapy.Curr Issues Mol Biol. 2023 Nov 25;45(12):9500-9525. doi: 10.3390/cimb45120595. Curr Issues Mol Biol. 2023. PMID: 38132441 Free PMC article. Review.
-
Exploring the Potential of MicroRNA Let-7c as a Therapeutic for Prostate Cancer.Mol Ther Nucleic Acids. 2019 Dec 6;18:927-937. doi: 10.1016/j.omtn.2019.09.031. Epub 2019 Oct 23. Mol Ther Nucleic Acids. 2019. PMID: 31760377 Free PMC article. Review.
Cited by
-
Antitumorigenic potential of Lactobacillus-derived extracellular vesicles: p53 succinylation and glycolytic reprogramming in intestinal epithelial cells via SIRT5 modulation.Cell Biol Toxicol. 2024 Aug 7;40(1):66. doi: 10.1007/s10565-024-09897-y. Cell Biol Toxicol. 2024. PMID: 39110260 Free PMC article.
-
Current Perspectives on Taxanes: Focus on Their Bioactivity, Delivery and Combination Therapy.Plants (Basel). 2021 Mar 17;10(3):569. doi: 10.3390/plants10030569. Plants (Basel). 2021. PMID: 33802861 Free PMC article. Review.
-
Identification of immune-related biomarkers for predicting neoadjuvant chemotherapy sensitivity in HER2 negative breast cancer via bioinformatics analysis.Gland Surg. 2022 Jun;11(6):1026-1036. doi: 10.21037/gs-22-234. Gland Surg. 2022. PMID: 35800743 Free PMC article.
-
Broadening the Horizons of RNA Delivery Strategies in Cancer Therapy.Bioengineering (Basel). 2022 Oct 19;9(10):576. doi: 10.3390/bioengineering9100576. Bioengineering (Basel). 2022. PMID: 36290544 Free PMC article. Review.
-
Photosensitive Nanoprobes for Rapid Isolation and Size-Specific Enrichment of Synthetic and Extracellular Vesicle Subpopulations.Adv Funct Mater. 2024 Aug 22;34(34):2400390. doi: 10.1002/adfm.202400390. Epub 2024 Mar 29. Adv Funct Mater. 2024. PMID: 39372670
References
LinkOut - more resources
Full Text Sources
Research Materials

