Different Families of Retrotransposons and DNA Transposons Are Actively Transcribed and May Have Transposed Recently in Physcomitrium (Physcomitrella) patens
- PMID: 32973835
- PMCID: PMC7466625
- DOI: 10.3389/fpls.2020.01274
Different Families of Retrotransposons and DNA Transposons Are Actively Transcribed and May Have Transposed Recently in Physcomitrium (Physcomitrella) patens
Abstract
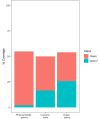
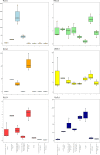
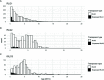
Similarly to other plant genomes of similar size, more than half of the genome of P. patens is covered by Transposable Elements (TEs). However, the composition and distribution of P. patens TEs is quite peculiar, with Long Terminal Repeat (LTR)-retrotransposons, which form patches of TE-rich regions interleaved with gene-rich regions, accounting for the vast majority of the TE space. We have already shown that RLG1, the most abundant TE in P. patens, is expressed in non-stressed protonema tissue. Here we present a non-targeted analysis of the TE expression based on RNA-Seq data and confirmed by qRT-PCR analyses that shows that, at least four LTR-RTs (RLG1, RLG2, RLC4 and tRLC5) and one DNA transposon (PpTc2) are expressed in P. patens. These TEs are expressed during development or under stresses that P. patens frequently faces, such as dehydratation/rehydratation stresses, suggesting that TEs have ample possibilities to transpose during P. patens life cycle. Indeed, an analysis of the TE polymorphisms among four different P. patens accessions shows that different TE families have recently transposed in this species and have generated genetic variability that may have phenotypic consequences, as a fraction of the TE polymorphisms are within or close to genes. Among the transcribed and mobile TEs, tRLC5 is particularly interesting as it concentrates in a single position per chromosome that could coincide with the centromere, and its expression is specifically induced in young sporophyte, where meiosis takes place.
Keywords: Physcomitrium (Physcomitrella) patens; centromere; genetic variability; transcription; transposable element.
Copyright © 2020 Vendrell-Mir, López-Obando, Nogué and Casacuberta.
Figures


Similar articles
-
The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution.Plant J. 2018 Feb;93(3):515-533. doi: 10.1111/tpj.13801. Plant J. 2018. PMID: 29237241
-
Mammalian transposable elements and their impacts on genome evolution.Chromosome Res. 2018 Mar;26(1-2):25-43. doi: 10.1007/s10577-017-9570-z. Epub 2018 Feb 1. Chromosome Res. 2018. PMID: 29392473 Free PMC article. Review.
-
New insights into nested long terminal repeat retrotransposons in Brassica species.Mol Plant. 2013 Mar;6(2):470-82. doi: 10.1093/mp/sss081. Epub 2012 Aug 28. Mol Plant. 2013. PMID: 22930733
-
Transposable elements generate regulatory novelty in a tissue-specific fashion.BMC Genomics. 2018 Jun 18;19(1):468. doi: 10.1186/s12864-018-4850-3. BMC Genomics. 2018. PMID: 29914366 Free PMC article.
-
Use of retrotransposon-derived genetic markers to analyse genomic variability in plants.Funct Plant Biol. 2018 Jan;46(1):15-29. doi: 10.1071/FP18098. Funct Plant Biol. 2018. PMID: 30939255 Review.
Cited by
-
Absence of major epigenetic and transcriptomic changes accompanying an interspecific cross between peach and almond.Hortic Res. 2022 May 26;9:uhac127. doi: 10.1093/hr/uhac127. eCollection 2022. Hortic Res. 2022. PMID: 35928404 Free PMC article.
-
Charting the genomic landscape of seed-free plants.Nat Plants. 2021 May;7(5):554-565. doi: 10.1038/s41477-021-00888-z. Epub 2021 Apr 5. Nat Plants. 2021. PMID: 33820965 Review.

