A Paradigm for Peptide Hormone-GPCR Analyses
- PMID: 32961885
- PMCID: PMC7570734
- DOI: 10.3390/molecules25184272
A Paradigm for Peptide Hormone-GPCR Analyses
Abstract
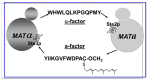
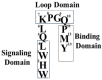
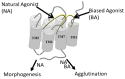
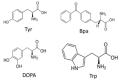
Work from our laboratories over the last 35 years that has focused on Ste2p, a G protein-coupled receptor (GPCR), and its tridecapeptide ligand α-factor is reviewed. Our work utilized the yeast Saccharomyces cerevisiae as a model system for understanding peptide-GPCR interactions. It explored the structure and function of synthetic α-factor analogs and biosynthetic receptor domains, as well as designed mutations of Ste2p. The results and conclusions are described using the nuclear magnetic resonance interrogation of synthetic Ste2p transmembrane domains (TMs), the fluorescence interrogation of agonist and antagonist binding, the biochemical crosslinking of peptide analogs to Ste2p, and the phenotypes of receptor mutants. We identified the ligand-binding domain in Ste2p, the functional assemblies of TMs, unexpected and interesting ligand analogs; gained insights into the bound α-factor structure; and unraveled the function and structures of various Ste2p domains, including the N-terminus, TMs, loops connecting the TMs, and the C-terminus. Our studies showed interactions between specific residues of Ste2p in an active state, but not resting state, and the effect of ligand activation on the dimerization of Ste2p. We show that, using a battery of different biochemical and genetic approaches, deep insight can be gained into the structure and conformational dynamics of GPCR-peptide interactions in the absence of a crystal structure.
Keywords: G protein-coupled receptors; Saccharomyces cerevisiae; chemical crosslinking; fluorescence screening; nuclear magnetic resonance; peptide analogs; peptide pheromone; photoactivated crosslinking; receptor mutation; receptor-ligand interaction.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Comparison of Experimental Approaches Used to Determine the Structure and Function of the Class D G Protein-Coupled Yeast α-Factor Receptor.Biomolecules. 2022 May 30;12(6):761. doi: 10.3390/biom12060761. Biomolecules. 2022. PMID: 35740886 Free PMC article. Review.
-
Novobiocin and peptide analogs of α-factor are positive allosteric modulators of the yeast G protein-coupled receptor Ste2p.Biochim Biophys Acta. 2015 Apr;1848(4):916-24. doi: 10.1016/j.bbamem.2014.12.024. Epub 2015 Jan 7. Biochim Biophys Acta. 2015. PMID: 25576192 Free PMC article.
-
The alpha-factor mating pheromone of Saccharomyces cerevisiae: a model for studying the interaction of peptide hormones and G protein-coupled receptors.Peptides. 2004 Sep;25(9):1441-63. doi: 10.1016/j.peptides.2003.11.028. Peptides. 2004. PMID: 15374647 Review.
-
Changes in conformation at the cytoplasmic ends of the fifth and sixth transmembrane helices of a yeast G protein-coupled receptor in response to ligand binding.Biochemistry. 2011 Aug 16;50(32):6841-54. doi: 10.1021/bi200254h. Epub 2011 Jul 12. Biochemistry. 2011. PMID: 21728340 Free PMC article.
-
Cross-linking of a DOPA-containing peptide ligand into its G protein-coupled receptor.Biochemistry. 2009 Mar 10;48(9):2033-44. doi: 10.1021/bi802061z. Biochemistry. 2009. PMID: 19152328 Free PMC article.
Cited by
-
Developing novel antifungals: lessons from G protein-coupled receptors.Trends Pharmacol Sci. 2023 Mar;44(3):162-174. doi: 10.1016/j.tips.2022.12.002. Trends Pharmacol Sci. 2023. PMID: 36801017 Free PMC article. Review.
-
ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.Elife. 2021 Nov 25;10:e72583. doi: 10.7554/eLife.72583. Elife. 2021. PMID: 34821552 Free PMC article.
-
Activation mechanism of the class D fungal GPCR dimer Ste2.Nature. 2022 Mar;603(7902):743-748. doi: 10.1038/s41586-022-04498-3. Epub 2022 Mar 16. Nature. 2022. PMID: 35296853 Free PMC article.
-
Comparison of Experimental Approaches Used to Determine the Structure and Function of the Class D G Protein-Coupled Yeast α-Factor Receptor.Biomolecules. 2022 May 30;12(6):761. doi: 10.3390/biom12060761. Biomolecules. 2022. PMID: 35740886 Free PMC article. Review.
-
Oligomerization of yeast α-factor receptor detected by fluorescent energy transfer between ligands.Biophys J. 2021 Nov 16;120(22):5090-5106. doi: 10.1016/j.bpj.2021.10.005. Epub 2021 Oct 8. Biophys J. 2021. PMID: 34627767 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

