Closely related reovirus lab strains induce opposite expression of RIG-I/IFN-dependent versus -independent host genes, via mechanisms of slow replication versus polymorphisms in dsRNA binding σ3 respectively
- PMID: 32956403
- PMCID: PMC7529228
- DOI: 10.1371/journal.ppat.1008803
Closely related reovirus lab strains induce opposite expression of RIG-I/IFN-dependent versus -independent host genes, via mechanisms of slow replication versus polymorphisms in dsRNA binding σ3 respectively
Abstract
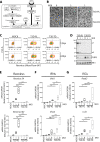
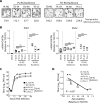
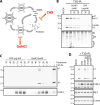
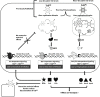
The Dearing isolate of Mammalian orthoreovirus (T3D) is a prominent model of virus-host relationships and a candidate oncolytic virotherapy. Closely related laboratory strains of T3D, originating from the same ancestral T3D isolate, were recently found to exhibit significantly different oncolytic properties. Specifically, the T3DPL strain had faster replication kinetics in a panel of cancer cells and improved tumor regression in an in vivo melanoma model, relative to T3DTD. In this study, we discover that T3DPL and T3DTD also differentially activate host signalling pathways and downstream gene transcription. At equivalent infectious dose, T3DTD induces higher IRF3 phosphorylation and expression of type I IFNs and IFN-stimulated genes (ISGs) than T3DPL. Using mono-reassortants with intermediate replication kinetics and pharmacological inhibitors of reovirus replication, IFN responses were found to inversely correlate with kinetics of virus replication. In other words, slow-replicating T3D strains induce more IFN signalling than fast-replicating T3D strains. Paradoxically, during co-infections by T3DPL and T3DTD, there was still high IRF3 phosphorylation indicating a phenodominant effect by the slow-replicating T3DTD. Using silencing and knock-out of RIG-I to impede IFN, we found that IFN induction does not affect the first round of reovirus replication but does prevent cell-cell spread in a paracrine fashion. Accordingly, during co-infections, T3DPL continues to replicate robustly despite activation of IFN by T3DTD. Using gene expression analysis, we discovered that reovirus can also induce a subset of genes in a RIG-I and IFN-independent manner; these genes were induced more by T3DPL than T3DTD. Polymorphisms in reovirus σ3 viral protein were found to control activation of RIG-I/ IFN-independent genes. Altogether, the study reveals that single amino acid polymorphisms in reovirus genomes can have large impact on host gene expression, by both changing replication kinetics and by modifying viral protein activity, such that two closely related T3D strains can induce opposite cytokine landscapes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Polymorphisms in the Most Oncolytic Reovirus Strain Confer Enhanced Cell Attachment, Transcription, and Single-Step Replication Kinetics.J Virol. 2020 Jan 31;94(4):e01937-19. doi: 10.1128/JVI.01937-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31776267 Free PMC article.
-
Single Amino Acid Differences between Closely Related Reovirus T3D Lab Strains Alter Oncolytic Potency In Vitro and In Vivo.J Virol. 2020 Jan 31;94(4):e01688-19. doi: 10.1128/JVI.01688-19. Print 2020 Jan 31. J Virol. 2020. PMID: 31748391 Free PMC article.
-
Reovirus Core Proteins λ1 and σ2 Promote Stability of Disassembly Intermediates and Influence Early Replication Events.J Virol. 2020 Aug 17;94(17):e00491-20. doi: 10.1128/JVI.00491-20. Print 2020 Aug 17. J Virol. 2020. PMID: 32581098 Free PMC article.
-
Reoviruses and the interferon system.Curr Top Microbiol Immunol. 1998;233(Pt 2):125-45. doi: 10.1007/978-3-642-72095-6_7. Curr Top Microbiol Immunol. 1998. PMID: 9599935 Review.
-
Modification of mammalian reoviruses for use as oncolytic agents.Expert Opin Biol Ther. 2009 Dec;9(12):1509-20. doi: 10.1517/14712590903307370. Expert Opin Biol Ther. 2009. PMID: 19916732 Review.
Cited by
-
U5 snRNP Core Proteins Are Key Components of the Defense Response against Viral Infection through Their Roles in Programmed Cell Death and Interferon Induction.Viruses. 2022 Dec 3;14(12):2710. doi: 10.3390/v14122710. Viruses. 2022. PMID: 36560714 Free PMC article.
-
Captivating Perplexities of Spinareovirinae 5' RNA Caps.Viruses. 2021 Feb 13;13(2):294. doi: 10.3390/v13020294. Viruses. 2021. PMID: 33668598 Free PMC article. Review.
-
Improved oncolytic activity of a reovirus mutant that displays enhanced virus spread due to reduced cell attachment.Mol Ther Oncolytics. 2023 Nov 2;31:100743. doi: 10.1016/j.omto.2023.100743. eCollection 2023 Dec 19. Mol Ther Oncolytics. 2023. PMID: 38033400 Free PMC article.
-
The reovirus μ2 C-terminal loop inversely regulates NTPase and transcription functions versus binding to factory-forming μNS and promotes replication in tumorigenic cells.J Virol. 2021 Apr 26;95(10):e02006-20. doi: 10.1128/JVI.02006-20. Epub 2021 Mar 3. J Virol. 2021. PMID: 33658345 Free PMC article.
-
The Reovirus σ3 Protein Inhibits NF-κB-Dependent Antiviral Signaling.J Virol. 2022 May 11;96(9):e0051522. doi: 10.1128/jvi.00515-22. Epub 2022 Apr 13. J Virol. 2022. PMID: 35416720 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

