Functional and structural characterization of allosteric activation of phospholipase Cε by Rap1A
- PMID: 32948655
- PMCID: PMC7864056
- DOI: 10.1074/jbc.RA120.015685
Functional and structural characterization of allosteric activation of phospholipase Cε by Rap1A
Abstract
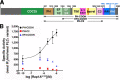
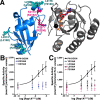
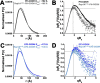
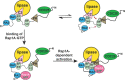
Phospholipase Cε (PLCε) is activated downstream of G protein-coupled receptors and receptor tyrosine kinases through direct interactions with small GTPases, including Rap1A and Ras. Although Ras has been reported to allosterically activate the lipase, it is not known whether Rap1A has the same ability or what its molecular mechanism might be. Rap1A activates PLCε in response to the stimulation of β-adrenergic receptors, translocating the complex to the perinuclear membrane. Because the C-terminal Ras association (RA2) domain of PLCε was proposed to the primary binding site for Rap1A, we first confirmed using purified proteins that the RA2 domain is indeed essential for activation by Rap1A. However, we also showed that the PLCε pleckstrin homology (PH) domain and first two EF hands (EF1/2) are required for Rap1A activation and identified hydrophobic residues on the surface of the RA2 domain that are also necessary. Small-angle X-ray scattering showed that Rap1A binding induces and stabilizes discrete conformational states in PLCε variants that can be activated by the GTPase. These data, together with the recent structure of a catalytically active fragment of PLCε, provide the first evidence that Rap1A, and by extension Ras, allosterically activate the lipase by promoting and stabilizing interactions between the RA2 domain and the PLCε core.
Keywords: G protein; Ras-related protein 1 (Rap1); calcium intracellular release; cardiovascular disease; cell signaling; conformational change; diacylglycerol; membrane enzyme; phosphatidylinositol signaling; phospholipase C; protein kinase C (PKC); small-angle X-ray scattering (SAXS); structural biology.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Similar articles
-
Structure and regulation of phospholipase Cβ and ε at the membrane.Chem Phys Lipids. 2021 Mar;235:105050. doi: 10.1016/j.chemphyslip.2021.105050. Epub 2021 Jan 7. Chem Phys Lipids. 2021. PMID: 33422547 Free PMC article. Review.
-
Role of the CDC25 homology domain of phospholipase Cepsilon in amplification of Rap1-dependent signaling.J Biol Chem. 2001 Aug 10;276(32):30301-7. doi: 10.1074/jbc.M103530200. Epub 2001 Jun 6. J Biol Chem. 2001. PMID: 11395506
-
Phospholipase Cepsilon guanine nucleotide exchange factor activity and activation of Rap1.Methods Enzymol. 2006;407:281-90. doi: 10.1016/S0076-6879(05)07024-2. Methods Enzymol. 2006. PMID: 16757332
-
Structure of phospholipase Cε reveals an integrated RA1 domain and previously unidentified regulatory elements.Commun Biol. 2020 Aug 14;3(1):445. doi: 10.1038/s42003-020-01178-8. Commun Biol. 2020. PMID: 32796910 Free PMC article.
-
PLCε mediated sustained signaling pathways.Adv Biol Regul. 2015 Jan;57:17-23. doi: 10.1016/j.jbior.2014.09.014. Epub 2014 Oct 5. Adv Biol Regul. 2015. PMID: 25453218 Free PMC article. Review.
Cited by
-
Small-angle X-ray scattering studies of enzymes.Curr Opin Chem Biol. 2023 Feb;72:102232. doi: 10.1016/j.cbpa.2022.102232. Epub 2022 Nov 30. Curr Opin Chem Biol. 2023. PMID: 36462455 Free PMC article. Review.
-
Soluble cyclase-mediated nuclear cAMP synthesis is sufficient for cell proliferation.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2208749120. doi: 10.1073/pnas.2208749120. Epub 2023 Jan 19. Proc Natl Acad Sci U S A. 2023. PMID: 36656863 Free PMC article.
-
Structure and regulation of phospholipase Cβ and ε at the membrane.Chem Phys Lipids. 2021 Mar;235:105050. doi: 10.1016/j.chemphyslip.2021.105050. Epub 2021 Jan 7. Chem Phys Lipids. 2021. PMID: 33422547 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

