Enzymatic degradation of RNA causes widespread protein aggregation in cell and tissue lysates
- PMID: 32945072
- PMCID: PMC7534620
- DOI: 10.15252/embr.201949585
Enzymatic degradation of RNA causes widespread protein aggregation in cell and tissue lysates
Abstract
Most proteins in cell and tissue lysates are soluble. We show here that in lysate from human neurons, more than 1,300 proteins are maintained in a soluble and functional state by association with endogenous RNA, as degradation of RNA invariably leads to protein aggregation. The majority of these proteins lack conventional RNA-binding domains. Using synthetic oligonucleotides, we identify the importance of nucleic acid structure, with single-stranded pyrimidine-rich bulges or loops surrounded by double-stranded regions being particularly efficient in the maintenance of protein solubility. These experiments also identify an apparent one-to-one protein-nucleic acid stoichiometry. Furthermore, we show that protein aggregates isolated from brain tissue from Amyotrophic Lateral Sclerosis patients can be rendered soluble after refolding by both RNA and synthetic oligonucleotides. Together, these findings open new avenues for understanding the mechanism behind protein aggregation and shed light on how certain proteins remain soluble.
Keywords: motor neurone disease; neurodegeneration; phase transition; protein precipitation; ribonuclease.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
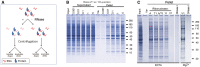
- A
Diagram showing the experimental design.
- B
SDS–PAGE analysis of soluble (Supernatant) and insoluble proteins (Pellet) from human neurons after treatment for one hour with a mixture of RNase A and RNase T1 (A/T1), or vehicle (Ve‐).
- C
Protein aggregation (Pellet) after incubation with different ribonucleases or DNase I in the presence of either EDTA or Mg2+. Ribonucleases used were RNase A (A), RNase T1 (T1), a mixture of RNase A and RNase T1 (A/T1), RNase 1f (1f) and RNase V1 (V1).
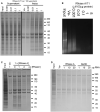
- A
SDS–PAGE analysis of soluble (Supernatant) and aggregated (Pellet) proteins after treatment of mouse brain tissue lysate with increasing amounts of a mixture of RNase A and T1 (A/T1) or vehicle (Ve−).
- B
Agarose gel electrophoresis analysis of RNA isolated from RNase‐treated human neuronal cell lysate.
- C, D
Insoluble proteins collected by centrifugation after co‐treatment of human neuronal cell lysate with RNase A and an RNase A inhibitor (RNasin, C), or following the addition of RNA pre‐hydrolysed by RNase A or alkaline hydrolysis (NaOH, D).
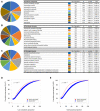
- A–C
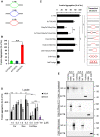
Top ten gene ontology classes by Cellular component (A), Molecular function (B), or Biological process (C).
- D, E
Cumulative distribution of the proportion of predicted low‐complexity regions (D) or unstructured regions (E) in the RNase‐aggregated proteins (Red) or random sets of proteins (Blue).

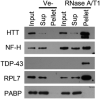
- A, B
Western blot analysis of soluble and aggregated proteins after RNase treatment of lysate from human neurons (A), or HEK293T cells expressing GFP‐Aβ or GFP (B).
- C
Effect of an RNase A inhibitor (RNasin) on RNase A mediated protein aggregation in neuronal cell lysate.

- A
Diagram showing the renaturing assay.
- B
Effect of RNA/protein ratio on protein aggregation after renaturing.
- C
Coomassie‐stained gel of soluble (Sup 1/2) and aggregated (Pel 1/2) proteins after removal of GuHCl in the presence (+) or absence (−) of total RNA. Asterisk (*) denotes added RNase A.
- D
As in (C) but analysed by Western blot.
- E
ATP‐hydrolysing activity of proteins renatured with total RNA or buffer (Ve−) after removal of GuHCl.
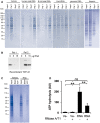
- A
Coomassie‐stained SDS‐PAGE gels of RNase‐aggregated proteins from human neurons renatured with various polyanions. Input represents 1/10th of each sample, taken before the first centrifugation, and Pel 1 and Sup 1 are the pellet and the supernatant, respectively, recovered after the first centrifugation. Pel 2 represents the pellet obtained after treating the Sup 1 fraction with RNase A/T1 followed by centrifugation. See experimental outline in Fig 3A for a full description.
- B
Renaturing of recombinant TDP‐43 using increasing amounts of total RNA. Pel 1 represents aggregated protein after renaturing. Pel 2 is the aggregated protein after the soluble fraction (Sup 1) has been treated with RNase A/T1.
- C
Gel electrophoresis analysis of proteins from human neurons renatured with vehicle or total RNA and then captured with ATP‐agarose beads (ATP). Input represents 1/10th of each sample taken directly after renaturation, before the first centrifugation, and Blank represents the sample without any ATP‐agarose beads present.
- D
ATP‐hydrolysing activity of RNase‐aggregated proteins from Jurkat T cells renatured in the presence (RNA) or absence (Ve−) of RNA.
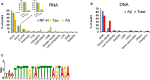
- A
Genomic attributes of RNA associated with renatured NF‐H, Tau or Aβ. Inset shows which fractions of the peaks that derive from exons or introns have their origin in coding or non‐coding transcripts, respectively.
- B
Characterisation of DNA associated with soluble, renatured Aβ or all proteins (Total), captured either by immunoprecipitation (Aβ) or absorption to membranes (Total).
- C
Sequence logo of the computationally identified motif (M1) in the solubilising genomic DNA of the Total samples.
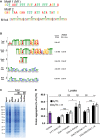
- A
Sequence of the selected M1 motif and a random control oligo (Motif 2, M2) investigated for their renaturing capacity. Both motifs were assembled into four consecutive repeats of the same motif (e.g. M1x4). The underlined region of M1 represents the computationally identified motif while the two extra 5′ nucleotides (GC) were added for cloning purposes.
- B
Alignment of motifs identified in the nucleic acids associated with soluble proteins after renaturation. All motifs are compared with the M1 motif identified in the Total DNA samples. The quoted original E‐value represents the value obtained in the MEME‐Chip analysis and the Similarity P‐value is the P‐value given by the TomTom motif comparison software package, compared with the Total DNA motif.
- C
Coomassie‐stained SDS‐PAGE gel of aggregated proteins (Pellet) following renaturation with either single‐stranded (For or Rev) or complementary (For/Rev) M1x4 or M2x4 DNA oligonucleotides. gDNA represents genomic DNA, and Ve‐ represents vehicle.
- D
Amount of protein aggregation observed after treating cell lysate from Jurkat T cells with various oligonucleotides and RNase A/T1 (A/T1) and, if indicated, together with RNase H (A/T1 + H).
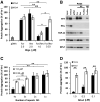
- A
Proportion of aggregated proteins after renaturation with either single‐stranded (Forward or Reverse) or complementary strands (For/Rev) of the selected M1x4 or the control oligonucleotide, M2x4.
- B
Western blot analysis of proteins renatured with either single‐stranded or double‐stranded M1x4 or M2x4 DNA oligonucleotides.
- C
The effect on protein re‐aggregation from varying the number of DNA motif repeats when complementary oligonucleotides are given together.
- D
Protein aggregation after renaturation with the M1x4 DNA oligonucleotides pre‐annealed with 100 mM NaCl or vehicle (None).
- A
Cartoons of ds/ss/ds oligonucleotides having either pyrimidines (T or C) or purines (A) in the ss region.
- B
Proportion of protein aggregation following renaturing with the ds/ss/ds oligonucleotides shown in (A).
- C
Renaturing capacity of structurally different oligonucleotides. The diagrams on the right show a theoretical structure of each oligonucleotide. All oligonucleotides, except the 3x‐loop and 3x‐bulges, contain a stretch of 30 Ts in the single‐stranded regions and the same sequences (15 nucleotides each) in the double‐stranded regions. The 3x‐loops and 3x‐bulges oligonucleotides have 3 stretches of 9 Ts and the same sequence in the ds‐regions.
- D
Proportion of protein aggregation in Jurkat cell lysate supplemented with various amounts and configurations of the M1x4 or M2x4 DNA oligonucleotides.
- E
Insoluble proteins from two ALS brain tissues were chemically denatured in guanidine hydrochloride and treated with either buffer (Vehicle), total RNA from Jurkat cells or the complementary strands of the M1x4 DNA oligonucleotides (M1 F/R) (Pellet 1, top panel). After removal of GuHCl, the soluble fraction from these samples was treated with RNase and any aggregated proteins (Pellet 2) analysed by Western blot (middle panel). Remaining supernatants were then treated with Benzonase to degrade any remaining nucleic acids and aggregated proteins collected by centrifugation and analysed by Western blot (bottom panel). Proteins aggregated by enzymatic RNA degradation in Jurkat cell lysates, which do not contain NF‐H, were used as a control.
Similar articles
-
The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight?Brain. 2019 May 1;142(5):1176-1194. doi: 10.1093/brain/awz078. Brain. 2019. PMID: 30938443 Free PMC article. Review.
-
Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration.Proc Natl Acad Sci U S A. 2013 May 7;110(19):7778-83. doi: 10.1073/pnas.1219643110. Epub 2013 Apr 3. Proc Natl Acad Sci U S A. 2013. PMID: 23553836 Free PMC article.
-
Protein aggregation in amyotrophic lateral sclerosis.Acta Neuropathol. 2013 Jun;125(6):777-94. doi: 10.1007/s00401-013-1125-6. Epub 2013 May 15. Acta Neuropathol. 2013. PMID: 23673820 Free PMC article. Review.
-
Loss of TDP-43 causes age-dependent progressive motor neuron degeneration.Brain. 2013 May;136(Pt 5):1371-82. doi: 10.1093/brain/awt029. Epub 2013 Feb 28. Brain. 2013. PMID: 23449777
-
Disease-linked TDP-43 hyperphosphorylation suppresses TDP-43 condensation and aggregation.EMBO J. 2022 Apr 19;41(8):e108443. doi: 10.15252/embj.2021108443. Epub 2022 Feb 3. EMBO J. 2022. PMID: 35112738 Free PMC article.
Cited by
-
Addressing Critical Issues Related to Storage and Stability of the Vault Nanoparticle Expressed and Purified from Komagataella phaffi.Int J Mol Sci. 2023 Feb 20;24(4):4214. doi: 10.3390/ijms24044214. Int J Mol Sci. 2023. PMID: 36835627 Free PMC article.
-
Disrupting pathologic phase transitions in neurodegeneration.J Clin Invest. 2023 Jul 3;133(13):e168549. doi: 10.1172/JCI168549. J Clin Invest. 2023. PMID: 37395272 Free PMC article. Review.
-
Synchronized motion of interface residues for evaluating protein-RNA complex binding affinity: Application to aptamer-mediated inhibition of TDP-43 aggregates.Protein Sci. 2024 Dec;33(12):e5201. doi: 10.1002/pro.5201. Protein Sci. 2024. PMID: 39548508 Free PMC article.
-
1-L Transcription in Prion Diseases.Int J Mol Sci. 2024 Sep 15;25(18):9961. doi: 10.3390/ijms25189961. Int J Mol Sci. 2024. PMID: 39337449 Free PMC article.
-
RNA-dependent proteome solubility maintenance in Escherichia coli lysates analysed by quantitative mass spectrometry: Proteomic characterization in terms of isoelectric point, structural disorder, functional hub, and chaperone network.RNA Biol. 2024 Jan;21(1):1-18. doi: 10.1080/15476286.2024.2315383. Epub 2024 Feb 15. RNA Biol. 2024. PMID: 38361426 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

