Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion
- PMID: 32942285
- PMCID: PMC7116727
- DOI: 10.1038/s41586-020-2772-0
Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion
Abstract
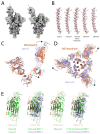
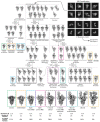
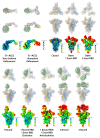
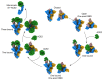
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is initiated by virus binding to the ACE2 cell-surface receptors1-4, followed by fusion of the virus and cell membranes to release the virus genome into the cell. Both receptor binding and membrane fusion activities are mediated by the virus spike glycoprotein5-7. As with other class-I membrane-fusion proteins, the spike protein is post-translationally cleaved, in this case by furin, into the S1 and S2 components that remain associated after cleavage8-10. Fusion activation after receptor binding is proposed to involve the exposure of a second proteolytic site (S2'), cleavage of which is required for the release of the fusion peptide11,12. Here we analyse the binding of ACE2 to the furin-cleaved form of the SARS-CoV-2 spike protein using cryo-electron microscopy. We classify ten different molecular species, including the unbound, closed spike trimer, the fully open ACE2-bound trimer and dissociated monomeric S1 bound to ACE2. The ten structures describe ACE2-binding events that destabilize the spike trimer, progressively opening up, and out, the individual S1 components. The opening process reduces S1 contacts and unshields the trimeric S2 core, priming the protein for fusion activation and dissociation of ACE2-bound S1 monomers. The structures also reveal refolding of an S1 subdomain after ACE2 binding that disrupts interactions with S2, which involves Asp61413-15 and leads to the destabilization of the structure of S2 proximal to the secondary (S2') cleavage site.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
SARS-CoV-2 spike engagement of ACE2 primes S2' site cleavage and fusion initiation.Proc Natl Acad Sci U S A. 2022 Jan 4;119(1):e2111199119. doi: 10.1073/pnas.2111199119. Proc Natl Acad Sci U S A. 2022. PMID: 34930824 Free PMC article.
-
Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2.PLoS Pathog. 2018 Aug 13;14(8):e1007236. doi: 10.1371/journal.ppat.1007236. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30102747 Free PMC article.
-
Proteolytic activation of SARS-CoV-2 spike protein.Microbiol Immunol. 2022 Jan;66(1):15-23. doi: 10.1111/1348-0421.12945. Epub 2021 Oct 12. Microbiol Immunol. 2022. PMID: 34561887 Free PMC article. Review.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
Cited by
-
Recent computational drug repositioning strategies against SARS-CoV-2.Comput Struct Biotechnol J. 2022;20:5713-5728. doi: 10.1016/j.csbj.2022.10.017. Epub 2022 Oct 17. Comput Struct Biotechnol J. 2022. PMID: 36277237 Free PMC article.
-
Non-uniform aspects of the SARS-CoV-2 intraspecies evolution reopen question of its origin.Int J Biol Macromol. 2022 Dec 1;222(Pt A):972-993. doi: 10.1016/j.ijbiomac.2022.09.184. Epub 2022 Sep 26. Int J Biol Macromol. 2022. PMID: 36174872 Free PMC article.
-
Potent SARS-CoV-2 neutralizing antibodies with therapeutic effects in two animal models.iScience. 2022 Dec 22;25(12):105596. doi: 10.1016/j.isci.2022.105596. Epub 2022 Nov 15. iScience. 2022. PMID: 36406861 Free PMC article.
-
Mutations in the B.1.1.7 SARS-CoV-2 Spike Protein Reduce Receptor-Binding Affinity and Induce a Flexible Link to the Fusion Peptide.Biomedicines. 2021 May 8;9(5):525. doi: 10.3390/biomedicines9050525. Biomedicines. 2021. PMID: 34066729 Free PMC article.
-
Immunoinformatic analysis of structural and epitope variations in the spike and Orf8 proteins of SARS-CoV-2/B.1.1.7.J Med Virol. 2021 Jul;93(7):4461-4468. doi: 10.1002/jmv.26931. Epub 2021 Mar 25. J Med Virol. 2021. PMID: 33704818 Free PMC article.
References
-
- Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

