Responsive fluorescent nucleotides serve as efficient substrates to probe terminal uridylyl transferase
- PMID: 32939524
- PMCID: PMC7611084
- DOI: 10.1039/d0cc05092j
Responsive fluorescent nucleotides serve as efficient substrates to probe terminal uridylyl transferase
Abstract
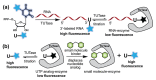
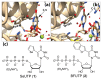
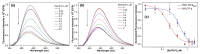
We repurposed a terminal uridylyl transferase enzyme to site-specifically label RNA with microenvironment sensing fluorescent nucleotide mimics, which in turn provided direct read-outs to estimate the binding affinities of the enzyme to RNA and nucleotide substrates. This enzyme-probe system provides insights into the catalytic cycle, and can facilitate the development of discovery platforms to identify robust enzyme inhibitors.
Conflict of interest statement
Conflicts of interest
The authors declare no conflict of interest.
Figures

Similar articles
-
Enzymatic Functionalization of RNA Oligonucleotides by Terminal Uridylyl Transferase Using Fluorescent and Clickable Nucleotide Analogs.Chem Asian J. 2024 Sep 16;19(18):e202400475. doi: 10.1002/asia.202400475. Epub 2024 Aug 19. Chem Asian J. 2024. PMID: 38949615
-
Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity.Nucleic Acids Res. 2015 Mar 11;43(5):2968-79. doi: 10.1093/nar/gkv122. Epub 2015 Feb 20. Nucleic Acids Res. 2015. PMID: 25712096 Free PMC article.
-
Elements of nucleotide specificity in the Trypanosoma brucei mitochondrial RNA editing enzyme RET2.J Chem Inf Model. 2012 May 25;52(5):1308-18. doi: 10.1021/ci3001327. Epub 2012 May 7. J Chem Inf Model. 2012. PMID: 22512810 Free PMC article.
-
Determinants of substrate specificity in RNA-dependent nucleotidyl transferases.Biochim Biophys Acta. 2008 Apr;1779(4):206-16. doi: 10.1016/j.bbagrm.2007.12.003. Epub 2007 Dec 14. Biochim Biophys Acta. 2008. PMID: 18177750 Free PMC article. Review.
-
RNA-specific ribonucleotidyl transferases.RNA. 2007 Nov;13(11):1834-49. doi: 10.1261/rna.652807. Epub 2007 Sep 13. RNA. 2007. PMID: 17872511 Free PMC article. Review.
Cited by
-
RNA Probes for Visualization of Sarcin/ricin Loop Depurination without Background Fluorescence.Chem Asian J. 2022 Dec 14;17(24):e202201077. doi: 10.1002/asia.202201077. Epub 2022 Nov 18. Chem Asian J. 2022. PMID: 36321802 Free PMC article.
-
Incorporation and Utility of a Responsive Ribonucleoside Analogue in Probing the Conformation of a Viral RNA Motif by Fluorescence and 19 F NMR Spectroscopy.Chembiochem. 2022 Feb 4;23(3):e202100601. doi: 10.1002/cbic.202100601. Epub 2021 Dec 7. Chembiochem. 2022. PMID: 34821449 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

