Elucidating the Basis for Permissivity of the MT-4 T-Cell Line to Replication of an HIV-1 Mutant Lacking the gp41 Cytoplasmic Tail
- PMID: 32938764
- PMCID: PMC7654282
- DOI: 10.1128/JVI.01334-20
Elucidating the Basis for Permissivity of the MT-4 T-Cell Line to Replication of an HIV-1 Mutant Lacking the gp41 Cytoplasmic Tail
Abstract
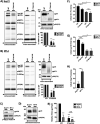
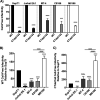
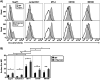
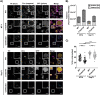
HIV-1 encodes an envelope glycoprotein (Env) that contains a long cytoplasmic tail (CT) harboring trafficking motifs implicated in Env incorporation into virus particles and viral transmission. In most physiologically relevant cell types, the gp41 CT is required for HIV-1 replication, but in the MT-4 T-cell line the gp41 CT is not required for a spreading infection. To help elucidate the role of the gp41 CT in HIV-1 transmission, in this study, we investigated the viral and cellular factors that contribute to the permissivity of MT-4 cells to gp41 CT truncation. We found that the kinetics of HIV-1 production and virus release are faster in MT-4 than in the other T-cell lines tested, but MT-4 cells express equivalent amounts of HIV-1 proteins on a per-cell basis relative to cells not permissive to CT truncation. MT-4 cells express higher levels of plasma-membrane-associated Env than nonpermissive cells, and Env internalization from the plasma membrane is less efficient than that from another T-cell line, SupT1. Paradoxically, despite the high levels of Env on the surface of MT-4 cells, 2-fold less Env is incorporated into virus particles produced from MT-4 than SupT1 cells. Contact-dependent transmission between cocultured 293T and MT-4 cells is higher than in cocultures of 293T with most other T-cell lines tested, indicating that MT-4 cells are highly susceptible to cell-to-cell infection. These data help to clarify the long-standing question of how MT-4 cells overcome the requirement for the HIV-1 gp41 CT and support a role for gp41 CT-dependent trafficking in Env incorporation and cell-to-cell transmission in physiologically relevant cell lines.IMPORTANCE The HIV-1 Env cytoplasmic tail (CT) is required for efficient Env incorporation into nascent particles and viral transmission in primary CD4+ T cells. The MT-4 T-cell line has been reported to support multiple rounds of infection of HIV-1 encoding a gp41 CT truncation. Uncovering the underlying mechanism of MT-4 T-cell line permissivity to gp41 CT truncation would provide key insights into the role of the gp41 CT in HIV-1 transmission. This study reveals that multiple factors contribute to the unique ability of a gp41 CT truncation mutant to spread in cultures of MT-4 cells. The lack of a requirement for the gp41 CT in MT-4 cells is associated with the combined effects of rapid HIV-1 protein production, high levels of cell-surface Env expression, and increased susceptibility to cell-to-cell transmission compared to nonpermissive cells.
Keywords: Env; HIV-1; HTLV-1; Tax; cytoplasmic tail; gp41; transmission; virological synapse.
Copyright © 2020 American Society for Microbiology.
Figures


Similar articles
-
Rab11-FIP1C Is Dispensable for HIV-1 Replication in Primary CD4+ T Cells, but Its Role Is Cell Type Dependent in Immortalized Human T-Cell Lines.J Virol. 2022 Dec 14;96(23):e0087622. doi: 10.1128/jvi.00876-22. Epub 2022 Nov 10. J Virol. 2022. PMID: 36354340 Free PMC article.
-
The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions.Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):343-8. doi: 10.1073/pnas.97.1.343. Proc Natl Acad Sci U S A. 2000. PMID: 10618420 Free PMC article.
-
Impact of natural polymorphism within the gp41 cytoplasmic tail of human immunodeficiency virus type 1 on the intracellular distribution of envelope glycoproteins and viral assembly.J Virol. 2007 Jan;81(1):125-40. doi: 10.1128/JVI.01659-06. Epub 2006 Oct 18. J Virol. 2007. PMID: 17050592 Free PMC article.
-
The frantic play of the concealed HIV envelope cytoplasmic tail.Retrovirology. 2013 May 24;10:54. doi: 10.1186/1742-4690-10-54. Retrovirology. 2013. PMID: 23705972 Free PMC article. Review.
-
HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation.J Mol Biol. 2011 Jul 22;410(4):582-608. doi: 10.1016/j.jmb.2011.04.042. J Mol Biol. 2011. PMID: 21762802 Free PMC article. Review.
Cited by
-
Rab11-FIP1C Is Dispensable for HIV-1 Replication in Primary CD4+ T Cells, but Its Role Is Cell Type Dependent in Immortalized Human T-Cell Lines.J Virol. 2022 Dec 14;96(23):e0087622. doi: 10.1128/jvi.00876-22. Epub 2022 Nov 10. J Virol. 2022. PMID: 36354340 Free PMC article.
-
The Assembly of HTLV-1-How Does It Differ from HIV-1?Viruses. 2024 Sep 27;16(10):1528. doi: 10.3390/v16101528. Viruses. 2024. PMID: 39459862 Free PMC article. Review.
-
Second site reversion of HIV-1 envelope protein baseplate mutations maps to the matrix protein.J Virol. 2024 Feb 20;98(2):e0174223. doi: 10.1128/jvi.01742-23. Epub 2024 Jan 9. J Virol. 2024. PMID: 38193694 Free PMC article.
-
YLMY Tyrosine Residue within the Cytoplasmic Tail of Newcastle Disease Virus Fusion Protein Regulates Its Surface Expression to Modulate Viral Budding and Pathogenicity.Microbiol Spectr. 2021 Dec 22;9(3):e0217321. doi: 10.1128/spectrum.02173-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34937182 Free PMC article.
-
A cGAS-mediated type I interferon response in human CD4+ T cells depends on productive infection and is conserved over HIV types and strains.J Virol. 2024 Oct 22;98(10):e0087724. doi: 10.1128/jvi.00877-24. Epub 2024 Sep 13. J Virol. 2024. PMID: 39269176
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

