Development and validation of a novel pseudogene pair-based prognostic signature for prediction of overall survival in patients with hepatocellular carcinoma
- PMID: 32938429
- PMCID: PMC7493157
- DOI: 10.1186/s12885-020-07391-2
Development and validation of a novel pseudogene pair-based prognostic signature for prediction of overall survival in patients with hepatocellular carcinoma
Abstract
Background: There is growing evidence that pseudogenes may serve as prognostic biomarkers in several cancers. The present study was designed to develop and validate an accurate and robust pseudogene pairs-based signature for the prognosis of hepatocellular carcinoma (HCC).
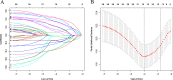
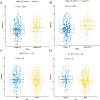
Methods: RNA-sequencing data from 374 HCC patients with clinical follow-up information were obtained from the Cancer Genome Atlas (TCGA) database and used in this study. Survival-related pseudogene pairs were identified, and a signature model was constructed by Cox regression analysis (univariate and least absolute shrinkage and selection operator). All individuals were classified into high- and low-risk groups based on the optimal cutoff. Subgroups analysis of the novel signature was conducted and validated in an independent cohort. Pearson correlation analyses were carried out between the included pseudogenes and the protein-coding genes based on their expression levels. Enrichment analysis was performed to predict the possible role of the pseudogenes identified in the signature.
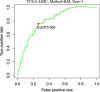
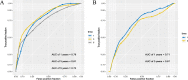
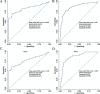
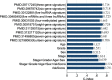
Results: A 19-pseudogene pair signature, which included 21 pseudogenes, was established. Patients in high-risk group demonstrated an increased the risk of adverse prognosis in the TCGA cohort and the external cohort (all P < 0.001). The novel pseudogene signature was independent of other conventional clinical variables used for survival prediction in HCC patients in the two cohorts revealed by the multivariate Cox regression analysis (all P < 0.001). Subgroup analysis further demonstrated the diagnostic value of the signature across different stages, grades, sexes, and age groups. The C-index of the prognostic signature was 0.761, which was not only higher than that of several previous risk models but was also much higher than that of a single age, sex, grade, and stage risk model. Furthermore, functional analysis revealed that the potential biological mechanisms mediated by these pseudogenes are primarily involved in cytokine receptor activity, T cell receptor signaling, chemokine signaling, NF-κB signaling, PD-L1 expression, and the PD-1 checkpoint pathway in cancer.
Conclusion: The novel proposed and validated pseudogene pair-based signature may serve as a valuable independent prognostic predictor for predicting survival of patients with HCC.
Keywords: Hepatocellular carcinoma; Pseudogene pairs; Signature; Survival.
Conflict of interest statement
None.
Figures

Similar articles
-
Six-long non-coding RNA signature predicts recurrence-free survival in hepatocellular carcinoma.World J Gastroenterol. 2019 Jan 14;25(2):220-232. doi: 10.3748/wjg.v25.i2.220. World J Gastroenterol. 2019. PMID: 30670911 Free PMC article.
-
Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma.World J Gastroenterol. 2018 Aug 14;24(30):3426-3439. doi: 10.3748/wjg.v24.i30.3426. World J Gastroenterol. 2018. PMID: 30122881 Free PMC article.
-
Identification and characterization of a 25-lncRNA prognostic signature for early recurrence in hepatocellular carcinoma.BMC Cancer. 2021 Oct 30;21(1):1165. doi: 10.1186/s12885-021-08827-z. BMC Cancer. 2021. PMID: 34717566 Free PMC article.
-
Mining Prognostic Biomarkers of Hepatocellular Carcinoma Based on Immune-Associated Genes.DNA Cell Biol. 2020 Apr;39(4):499-512. doi: 10.1089/dna.2019.5099. Epub 2020 Feb 18. DNA Cell Biol. 2020. PMID: 32069130 Review.
-
Pseudogene Transcripts in Head and Neck Cancer: Literature Review and In Silico Analysis.Genes (Basel). 2021 Aug 17;12(8):1254. doi: 10.3390/genes12081254. Genes (Basel). 2021. PMID: 34440428 Free PMC article. Review.
Cited by
-
Identification and validation of an epithelial mesenchymal transition-related gene pairs signature for prediction of overall survival in patients with skin cutaneous melanoma.PeerJ. 2022 Jan 21;10:e12646. doi: 10.7717/peerj.12646. eCollection 2022. PeerJ. 2022. PMID: 35116193 Free PMC article.
-
Identification of cuproptosis and ferroptosis-related subgroups and development of a signature for predicting prognosis and tumor microenvironment landscape in hepatocellular carcinoma.Transl Cancer Res. 2023 Dec 31;12(12):3327-3345. doi: 10.21037/tcr-23-685. Epub 2023 Dec 27. Transl Cancer Res. 2023. PMID: 38192999 Free PMC article.
-
Golgi-apparatus genes related signature for predicting the progression-free interval of patients with papillary thyroid carcinoma.BMC Med Genomics. 2023 Mar 27;16(1):60. doi: 10.1186/s12920-023-01485-z. BMC Med Genomics. 2023. PMID: 36973751 Free PMC article.
-
The World of Pseudogenes: New Diagnostic and Therapeutic Targets in Cancers or Still Mystery Molecules?Life (Basel). 2021 Dec 7;11(12):1354. doi: 10.3390/life11121354. Life (Basel). 2021. PMID: 34947885 Free PMC article. Review.
-
Ovarian Cancer Risk Scores Based on Immune-Related Pseudogenes to Predict Overall Survival and Guide Immunotherapy and Chemotherapy.Evid Based Complement Alternat Med. 2021 Oct 8;2021:1586312. doi: 10.1155/2021/1586312. eCollection 2021. Evid Based Complement Alternat Med. 2021. Retraction in: Evid Based Complement Alternat Med. 2023 Jun 21;2023:9819628. doi: 10.1155/2023/9819628 PMID: 34659427 Free PMC article. Retracted.
References
-
- Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2012;379(9822):1245–1255. - PubMed
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. - PubMed
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. - PubMed
-
- Villanueva A, Hernandez-Gea V, Llovet JM. Medical therapies for hepatocellular carcinoma: a critical view of the evidence. Nat Rev Gastroenterol Hepatol. 2013;10(1):34–42. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

