Similar patterns of genetic diversity and linkage disequilibrium in Western chimpanzees (Pan troglodytes verus) and humans indicate highly conserved mechanisms of MHC molecular evolution
- PMID: 32933484
- PMCID: PMC7491122
- DOI: 10.1186/s12862-020-01669-6
Similar patterns of genetic diversity and linkage disequilibrium in Western chimpanzees (Pan troglodytes verus) and humans indicate highly conserved mechanisms of MHC molecular evolution
Abstract
Background: Many species are threatened with extinction as their population sizes decrease with changing environments or face novel pathogenic threats. A reduction of genetic diversity at major histocompatibility complex (MHC) genes may have dramatic effects on populations' survival, as these genes play a key role in adaptive immunity. This might be the case for chimpanzees, the MHC genes of which reveal signatures of an ancient selective sweep likely due to a viral epidemic that reduced their population size a few million years ago. To better assess how this past event affected MHC variation in chimpanzees compared to humans, we analysed several indexes of genetic diversity and linkage disequilibrium across seven MHC genes on four cohorts of chimpanzees and we compared them to those estimated at orthologous HLA genes in a large set of human populations.
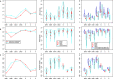
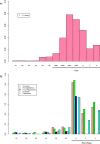
Results: Interestingly, the analyses uncovered similar patterns of both molecular diversity and linkage disequilibrium across the seven MHC genes in chimpanzees and humans. Indeed, in both species the greatest allelic richness and heterozygosity were found at loci A, B, C and DRB1, the greatest nucleotide diversity at loci DRB1, DQA1 and DQB1, and both significant global linkage disequilibrium and the greatest proportions of haplotypes in linkage disequilibrium were observed at pairs DQA1 ~ DQB1, DQA1 ~ DRB1, DQB1 ~ DRB1 and B ~ C. Our results also showed that, despite some differences among loci, the levels of genetic diversity and linkage disequilibrium observed in contemporary chimpanzees were globally similar to those estimated in small isolated human populations, in contrast to significant differences compared to large populations.
Conclusions: We conclude, first, that highly conserved mechanisms shaped the diversity of orthologous MHC genes in chimpanzees and humans. Furthermore, our findings support the hypothesis that an ancient demographic decline affecting the chimpanzee populations - like that ascribed to a viral epidemic - exerted a substantial effect on the molecular diversity of their MHC genes, albeit not more pronounced than that experienced by HLA genes in human populations that underwent rapid genetic drift during humans' peopling history. We thus propose a model where chimpanzees' MHC genes regenerated molecular variation through recombination/gene conversion and/or balancing selection after the selective sweep.
Keywords: Balancing selection; Demographic history; HLA; Human populations; Linkage disequilibrium; MHC; Nucleotide diversity; Patr; Population bottleneck; Selective sweep; Western chimpanzees.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
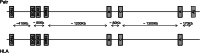

Similar articles
-
Mhc-DRB diversity of the chimpanzee (Pan troglodytes).Immunogenetics. 1992;37(1):1-11. doi: 10.1007/BF00223539. Immunogenetics. 1992. PMID: 1428007
-
Evidence for a complex demographic history of chimpanzees.Mol Biol Evol. 2004 May;21(5):799-808. doi: 10.1093/molbev/msh083. Epub 2004 Feb 12. Mol Biol Evol. 2004. PMID: 14963091
-
Polymorphism, recombination, and linkage disequilibrium within the HLA class II region.J Immunol. 1992 Jan 1;148(1):249-58. J Immunol. 1992. PMID: 1727870
-
The molecular signature of selection underlying human adaptations.Am J Phys Anthropol. 2006;Suppl 43:89-130. doi: 10.1002/ajpa.20518. Am J Phys Anthropol. 2006. PMID: 17103426 Review.
-
Distribution of HLA haplotypes across Japanese Archipelago: similarity, difference and admixture.J Hum Genet. 2015 Nov;60(11):683-90. doi: 10.1038/jhg.2015.90. Epub 2015 Jul 23. J Hum Genet. 2015. PMID: 26202576 Review.
Cited by
-
Balancing selection shapes population differentiation of major histocompatibility complex genes in wild golden snub-nosed monkeys.Curr Zool. 2023 Sep 24;70(5):596-606. doi: 10.1093/cz/zoad043. eCollection 2024 Oct. Curr Zool. 2023. PMID: 39463695 Free PMC article.
-
Like Wings of a Bird: Functional Divergence and Complementarity between HLA-A and HLA-B Molecules.Mol Biol Evol. 2021 Apr 13;38(4):1580-1594. doi: 10.1093/molbev/msaa325. Mol Biol Evol. 2021. PMID: 33320202 Free PMC article.
References
-
- Parham P. The immune system. 4th ed. New York: Garland Science; 2015.
-
- Lawlor DA, Ward FE, Ennis PD, Jackson AP, Parham P. HLA-A and B polymorphisms predate the divergence of humans and chimpanzees. Nature. 1988;335(6187):268–271. - PubMed
-
- Klein J, Bontrop RE, Dawkins RL, et al. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31(4):217–9. - PubMed
-
- Bontrop RE, Otting N, de Groot NG, Doxiadis GG. Major histocompatibility complex class II polymorphisms in primates. Immunol Rev. 1999;167:339–350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

