Genomic epidemiology and evolutionary dynamics of respiratory syncytial virus group B in Kilifi, Kenya, 2015-17
- PMID: 32913665
- PMCID: PMC7474930
- DOI: 10.1093/ve/veaa050
Genomic epidemiology and evolutionary dynamics of respiratory syncytial virus group B in Kilifi, Kenya, 2015-17
Abstract
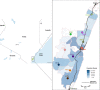
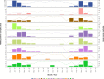
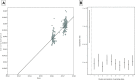
Respiratory syncytial virus (RSV) circulates worldwide, occurring seasonally in communities, and is a leading cause of acute respiratory illness in young children. There is paucity of genomic data from purposively sampled populations by which to investigate evolutionary dynamics and transmission patterns of RSV. Here we present an analysis of 295 RSV group B (RSVB) genomes from Kilifi, coastal Kenya, sampled from individuals seeking outpatient care in nine health facilities across a defined geographical area (∼890 km2), over two RSV epidemics between 2015 and 2017. RSVB diversity was characterized by multiple virus introductions into the area and co-circulation of distinct genetic clusters, which transmitted and diversified locally with varying frequency. Increase in relative genetic diversity paralleled seasonal virus incidence. Importantly, we identified a cluster of viruses that emerged in the 2016/17 epidemic, carrying distinct amino-acid signatures including a novel nonsynonymous change (K68Q) in antigenic site ∅ in the Fusion protein. RSVB diversity was additionally marked by signature nonsynonymous substitutions that were unique to particular genomic clusters, some under diversifying selection. Our findings provide insights into recent evolutionary and epidemiological behaviors of RSVB, and highlight possible emergence of a novel antigenic variant, which has implications on current prophylactic strategies in development.
Keywords: RSV; community; emergence; evolution; genomes.
© The Author(s) 2020. Published by Oxford University Press.
Figures

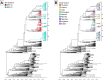
Similar articles
-
Successive Respiratory Syncytial Virus Epidemics in Local Populations Arise from Multiple Variant Introductions, Providing Insights into Virus Persistence.J Virol. 2015 Nov;89(22):11630-42. doi: 10.1128/JVI.01972-15. Epub 2015 Sep 9. J Virol. 2015. PMID: 26355091 Free PMC article.
-
The genomic evolutionary dynamics and global circulation patterns of respiratory syncytial virus.Nat Commun. 2024 Apr 10;15(1):3083. doi: 10.1038/s41467-024-47118-6. Nat Commun. 2024. PMID: 38600104 Free PMC article.
-
Whole genome analysis of local Kenyan and global sequences unravels the epidemiological and molecular evolutionary dynamics of RSV genotype ON1 strains.Virus Evol. 2018 Sep 24;4(2):vey027. doi: 10.1093/ve/vey027. eCollection 2018 Jul. Virus Evol. 2018. PMID: 30271623 Free PMC article.
-
Respiratory Syncytial Virus: The Influence of Serotype and Genotype Variability on Clinical Course of Infection.Int J Mol Sci. 2017 Aug 6;18(8):1717. doi: 10.3390/ijms18081717. Int J Mol Sci. 2017. PMID: 28783078 Free PMC article. Review.
-
Mucosal vaccines against respiratory syncytial virus.Curr Opin Virol. 2014 Jun;6:78-84. doi: 10.1016/j.coviro.2014.03.009. Epub 2014 Apr 29. Curr Opin Virol. 2014. PMID: 24794644 Free PMC article. Review.
Cited by
-
Molecular epidemiology of respiratory syncytial virus in children with acute respiratory illnesses in Africa: A systematic review and meta-analysis.J Glob Health. 2023 Jan 14;13:04001. doi: 10.7189/jogh.13.04001. J Glob Health. 2023. PMID: 36637855 Free PMC article.
-
Evolutionary dynamics of group A and B respiratory syncytial virus in China, 2009-2018.Arch Virol. 2021 Sep;166(9):2407-2418. doi: 10.1007/s00705-021-05139-2. Epub 2021 Jun 15. Arch Virol. 2021. PMID: 34131849
-
A systematic review on global RSV genetic data: Identification of knowledge gaps.Rev Med Virol. 2022 May;32(3):e2284. doi: 10.1002/rmv.2284. Epub 2021 Sep 20. Rev Med Virol. 2022. PMID: 34543489 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

