Involvement of the NF-κB signaling pathway in proliferation and invasion inhibited by Zwint-1 deficiency in Pancreatic Cancer Cells
- PMID: 32913455
- PMCID: PMC7477444
- DOI: 10.7150/jca.46173
Involvement of the NF-κB signaling pathway in proliferation and invasion inhibited by Zwint-1 deficiency in Pancreatic Cancer Cells
Abstract
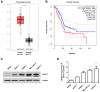
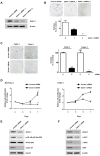
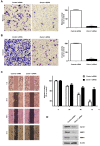
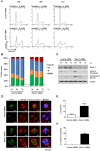
Pancreatic cancer (PC) is an intractable cancer that is difficult to diagnose early and has a 5-year survival rate of less than 8%. ZW10-interacting kinetochore protein (ZWINT) is a crucial gene that contributes to chromosome instability and is essential for spindle assembly and kinetochore-microtubule attachment during meiosis and mitosis. However, the mechanism through which Zwint-1 promotes PC progression is yet to be elucidated. Here, we report that Zwint-1 is highly expressed in clinical PC specimens (based on analysis of the Gene Expression Profiling Interactive Analysis database) and various PC cell lines. Importantly, Zwint-1-deficient PC cells showed reduced nuclear factor-kappa B (NF-κB) (Ser536) phosphorylation along with inhibited proliferation and colony formation due to downregulation of NF-κB-regulated genes such as CCND1, cIAP1/2, and XIAP. In addition, Zwint-1-deficient PC cells showed reduced invasion and migration abilities, and decreased expression levels of the metalloproteinases MMP2 and MMP9. Furthermore, Zwint-1 deficiency arrested the PC cell cycle at the G2/M phase because the chromosomes failed to segregate properly, and the apoptosis rate in these cells gradually increased, accompanied by increased caspase-3 activation and anti-poly (ADP ribose) polymerase cleavage. Apoptosis caused by Zwint-1 deficiency was demonstrated to occur through caspase-dependent pathways based on experiments involving treatment with a pan-caspase inhibitor (Z-VAD-Fmk). Thus, Zwint-1 contributes to cell growth, invasion, and survival through NF-κB signaling pathways, suggesting that it could serve as a PC biomarker and new therapeutic target.
Keywords: CCAN; KMN; NF-κB; RZZ; Zwint-1; cancer biology; pancreatic cancer.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
Hypoxia-Induced ZWINT Mediates Pancreatic Cancer Proliferation by Interacting With p53/p21.Front Cell Dev Biol. 2021 Nov 24;9:682131. doi: 10.3389/fcell.2021.682131. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34900978 Free PMC article.
-
A multidimensional analysis of ZW10 interacting kinetochore protein in human tumors.Am J Cancer Res. 2024 Jan 25;14(1):390-402. doi: 10.62347/MDPI5698. eCollection 2024. Am J Cancer Res. 2024. PMID: 38323280 Free PMC article.
-
Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control.Oncogene. 2006 Nov 2;25(52):6901-14. doi: 10.1038/sj.onc.1209687. Epub 2006 May 29. Oncogene. 2006. PMID: 16732327
-
ZWINT: A potential therapeutic biomarker in patients with glioblastoma correlates with cell proliferation and invasion.Oncol Rep. 2020 Jun;43(6):1831-1844. doi: 10.3892/or.2020.7573. Epub 2020 Apr 2. Oncol Rep. 2020. PMID: 32323832 Free PMC article.
-
ZWINT promotes the proliferation, migration, and invasion of cervical cancer cells by regulating the p53/p21 signaling pathway.Chin J Physiol. 2023 Sep-Oct;66(5):372-378. doi: 10.4103/cjop.CJOP-D-23-00001. Chin J Physiol. 2023. PMID: 37929349
Cited by
-
Prognostic and Clinicopathological Value of ZWINT Expression Levels in Patients with Lung Adenocarcinoma: A Systematic Review and Meta-analysis.Clinics (Sao Paulo). 2021 Nov 26;76:e3222. doi: 10.6061/clinics/2021/e3222. eCollection 2021. Clinics (Sao Paulo). 2021. PMID: 34852139 Free PMC article.
-
Identification of the cell cycle characteristics of non-small cell lung cancer and its relationship with tumor immune microenvironment, cell death pathways, and metabolic reprogramming.Front Endocrinol (Lausanne). 2023 Apr 6;14:1147366. doi: 10.3389/fendo.2023.1147366. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37091844 Free PMC article.
-
The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma.Cells. 2023 Mar 28;12(7):1029. doi: 10.3390/cells12071029. Cells. 2023. PMID: 37048102 Free PMC article.
-
Contributions of NFKB1 -94insertion/deletion ATTG polymorphism to the susceptibility of gastrointestinal cancers: A meta-analysis.J Cell Mol Med. 2021 Nov;25(22):10674-10683. doi: 10.1111/jcmm.17004. Epub 2021 Oct 21. J Cell Mol Med. 2021. PMID: 34672421 Free PMC article.
-
Hypoxia-Induced ZWINT Mediates Pancreatic Cancer Proliferation by Interacting With p53/p21.Front Cell Dev Biol. 2021 Nov 24;9:682131. doi: 10.3389/fcell.2021.682131. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34900978 Free PMC article.
References
-
- Westhorpe FG, Straight AF. Chromosome Segregation: Reconstituting the Kinetochore. Current biology: CB. 2016;26:R1242–r5. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

