High accuracy gene expression profiling of sorted cell subpopulations from breast cancer PDX model tissue
- PMID: 32911489
- PMCID: PMC7482927
- DOI: 10.1371/journal.pone.0238594
High accuracy gene expression profiling of sorted cell subpopulations from breast cancer PDX model tissue
Abstract
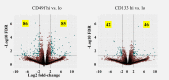
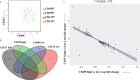
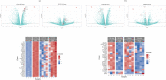
Intratumor Heterogeneity (ITH) is a functionally important property of tumor tissue and may be involved in drug resistance mechanisms. Although descriptions of ITH can be traced back to very early reports about cancer tissue, mechanistic investigations are still limited by the precision of analysis methods and access to relevant tissue sources. PDX models have provided a reproducible source of tissue with at least a partial representation of naturally occurring ITH. We investigated the properties of phenotypically distinct cell populations by Fluorescence activated cell sorting (FACS) tissue derived cells from multiple tumors from a triple negative breast cancer patient derived xenograft (PDX) model. We subsequently subjected each population to in depth gene expression analysis. Our findings suggest that process related gene expression changes (caused by tissue dissociation and FACS sorting) are restricted to Immediate Early Genes (IEGs). This allowed us to discover highly reproducible gene expression profiles of distinct cellular compartments identifiable by cell surface markers in this particular tumor model. Within the context of data from a previously published model our work suggests that gene expression profiles associated with hypoxia, stemness and drug resistance may reside in tumor subpopulations predictably growing in PDX models. This approach provides a novel opportunity for prospective mechanistic studies of ITH.
Conflict of interest statement
WP, ES, FT, WSD and RB are employees of BD Technologies and Innovation and FH and MF were employees of BD Technologies and Innovation at the time the work was carried out. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures


Similar articles
-
Immunophenotyping and Transcriptomic Outcomes in PDX-Derived TNBC Tissue.Mol Cancer Res. 2017 Apr;15(4):429-438. doi: 10.1158/1541-7786.MCR-16-0286-T. Epub 2016 Dec 30. Mol Cancer Res. 2017. PMID: 28039356
-
A large collection of integrated genomically characterized patient-derived xenografts highlighting the heterogeneity of triple-negative breast cancer.Int J Cancer. 2019 Oct 1;145(7):1902-1912. doi: 10.1002/ijc.32266. Epub 2019 Apr 4. Int J Cancer. 2019. PMID: 30859564
-
p53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancer.Breast Cancer Res. 2016 Jan 27;18(1):13. doi: 10.1186/s13058-016-0673-9. Breast Cancer Res. 2016. PMID: 26818199 Free PMC article.
-
Drug resistance profiling of a new triple negative breast cancer patient-derived xenograft model.BMC Cancer. 2019 Mar 7;19(1):205. doi: 10.1186/s12885-019-5401-2. BMC Cancer. 2019. PMID: 30845999 Free PMC article.
-
Establishment of chemosensitivity tests in triple-negative and BRCA-mutated breast cancer patient-derived xenograft models.PLoS One. 2019 Dec 10;14(12):e0225082. doi: 10.1371/journal.pone.0225082. eCollection 2019. PLoS One. 2019. PMID: 31821346 Free PMC article.
Cited by
-
Life history dynamics of evolving tumors: insights into task specialization, trade-offs, and tumor heterogeneity.Cancer Cell Int. 2024 Nov 6;24(1):364. doi: 10.1186/s12935-024-03538-4. Cancer Cell Int. 2024. PMID: 39506763 Free PMC article. Review.
-
Pterostilbene Changes Epigenetic Marks at Enhancer Regions of Oncogenes in Breast Cancer Cells.Antioxidants (Basel). 2021 Jul 30;10(8):1232. doi: 10.3390/antiox10081232. Antioxidants (Basel). 2021. PMID: 34439480 Free PMC article.
References
-
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12(5):323–34. - PubMed
-
- Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44(6):2259–65. Epub 1984/06/01. . - PubMed
-
- Heppner GH, Shekhar M. Tumor heterogeneity is fundamental to the tumor ecosystem. Oncology (Williston Park, NY). 2014;28(9):780–1. Epub 2014/09/17. . - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

