Interplay between Viscoelasticity and Force Rate Affects Sequential Unfolding in Polyproteins Pulled at Constant Velocity
- PMID: 32905266
- PMCID: PMC7467765
- DOI: 10.1021/acs.macromol.0c00278
Interplay between Viscoelasticity and Force Rate Affects Sequential Unfolding in Polyproteins Pulled at Constant Velocity
Abstract
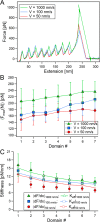
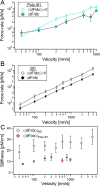
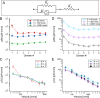
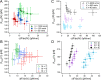
Polyproteins are unique constructs, comprised of folded protein domains in tandem and polymeric linkers. These macromolecules perform under biological stresses by modulating their response through partial unfolding and extending. Although these unfolding events are considered independent, a history dependence of forced unfolding within polyproteins was reported. Here we measure the unfolding of single poly(I91) octamers, complemented with Brownian dynamics simulations, displaying increasing hierarchy in unfolding-foces, accompanied by a decrease in the effective stiffness. This counters the existing understanding that relates stiffness with variations in domain size and probe stiffness, which is expected to reduce the unfolding forces with every consecutive unfolding event. We utilize a simple mechanistic viscoelastic model to show that two effects are combined within a sequential forced unfolding process: the viscoelastic properties of the growing linker chain lead to a hierarchy of the unfolding events, and force-rate application governs the unfolding kinetics.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Nonexponential kinetics captured in sequential unfolding of polyproteins over a range of loads.Curr Res Struct Biol. 2022 Apr 28;4:106-117. doi: 10.1016/j.crstbi.2022.04.003. eCollection 2022. Curr Res Struct Biol. 2022. PMID: 35540955 Free PMC article.
-
Correlations within polyprotein forced unfolding dwell-times introduce sequential dependency.J Struct Biol. 2020 Jun 1;210(3):107495. doi: 10.1016/j.jsb.2020.107495. Epub 2020 Mar 12. J Struct Biol. 2020. PMID: 32173465
-
Segmentation and the Entropic Elasticity of Modular Proteins.J Phys Chem Lett. 2018 Aug 16;9(16):4707-4713. doi: 10.1021/acs.jpclett.8b01925. Epub 2018 Aug 6. J Phys Chem Lett. 2018. PMID: 30058807
-
Interdomain linkers tailor the stability of immunoglobulin repeats in polyproteins.Biochem Biophys Res Commun. 2021 Apr 23;550:43-48. doi: 10.1016/j.bbrc.2021.02.114. Epub 2021 Mar 5. Biochem Biophys Res Commun. 2021. PMID: 33684619
-
The physics of pulling polyproteins: a review of single molecule force spectroscopy using the AFM to study protein unfolding.Rep Prog Phys. 2016 Jul;79(7):076601. doi: 10.1088/0034-4885/79/7/076601. Epub 2016 Jun 16. Rep Prog Phys. 2016. PMID: 27309041 Review.
Cited by
-
Nonexponential kinetics captured in sequential unfolding of polyproteins over a range of loads.Curr Res Struct Biol. 2022 Apr 28;4:106-117. doi: 10.1016/j.crstbi.2022.04.003. eCollection 2022. Curr Res Struct Biol. 2022. PMID: 35540955 Free PMC article.
References
LinkOut - more resources
Full Text Sources
