Conserved ESX-1 Substrates EspE and EspF Are Virulence Factors That Regulate Gene Expression
- PMID: 32900815
- PMCID: PMC7671884
- DOI: 10.1128/IAI.00289-20
Conserved ESX-1 Substrates EspE and EspF Are Virulence Factors That Regulate Gene Expression
Abstract
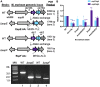
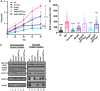
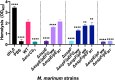
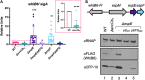
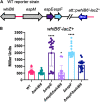
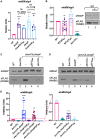
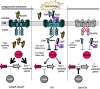
Mycobacterium tuberculosis, the cause of human tuberculosis, and Mycobacterium marinum, a nontubercular pathogen with a broad host range, require the ESX-1 secretion system for virulence. The ESX-1 system secretes proteins which cause phagosomal lysis within the macrophage via an unknown mechanism. As reported elsewhere (R. E. Bosserman et al., Proc Natl Acad Sci U S A 114:E10772-E10781, 2017, https://doi.org/10.1073/pnas.1710167114), we recently discovered that the ESX-1 system regulates gene expression in M. marinum This finding was confirmed in M. tuberculosis in reports by C. Sala et al. (PLoS Pathog 14:e1007491, 2018, https://doi.org/10.1371/journal.ppat.1007491) and A. M. Abdallah et al. (PLoS One 14:e0211003, 2019, https://doi.org/10.1371/journal.pone.0211003). We further demonstrated that a feedback control mechanism connects protein secretion to WhiB6-dependent expression of the esx-1 genes via an unknown mechanism. Here, we connect protein secretion and gene expression by showing for the first time that specific ESX-1 substrates have dual functions inside and outside the mycobacterial cell. We demonstrate that the EspE and EspF substrates negatively control esx-1 gene expression in the M. marinum cytoplasm through the conserved WhiB6 transcription factor. We found that EspE and EspF are required for virulence and promote lytic activity independently of the major EsxA and EsxB substrates. We show that the dual functions of EspE and EspF are conserved in the orthologous proteins from M. tuberculosis Our findings support a role for EspE and EspF in virulence that is independent of the EsxA and EsxB substrates and demonstrate that ESX-1 substrates have a conserved role in regulating gene expression.
Keywords: ESAT-6; ESX-1; Mycobacterium marinum; WhiB6; mycobacterial pathogenesis; protein secretion; type VII.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
EspM Is a Conserved Transcription Factor That Regulates Gene Expression in Response to the ESX-1 System.mBio. 2020 Feb 4;11(1):e02807-19. doi: 10.1128/mBio.02807-19. mBio. 2020. PMID: 32019792 Free PMC article.
-
EspH is a hypervirulence factor for Mycobacterium marinum and essential for the secretion of the ESX-1 substrates EspE and EspF.PLoS Pathog. 2018 Aug 13;14(8):e1007247. doi: 10.1371/journal.ppat.1007247. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30102741 Free PMC article.
-
The regulatory functions of ESX-1 substrates, EspE and EspF, are separable from secretion.J Bacteriol. 2024 Sep 19;206(9):e0027124. doi: 10.1128/jb.00271-24. Epub 2024 Aug 13. J Bacteriol. 2024. PMID: 39136451 Free PMC article.
-
Modeling Tubercular ESX-1 Secretion Using Mycobacterium marinum.Microbiol Mol Biol Rev. 2020 Sep 2;84(4):e00082-19. doi: 10.1128/MMBR.00082-19. Print 2020 Nov 18. Microbiol Mol Biol Rev. 2020. PMID: 32878966 Free PMC article. Review.
-
ESX/Type VII Secretion Systems-An Important Way Out for Mycobacterial Proteins.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.psib-0029-2019. doi: 10.1128/microbiolspec.PSIB-0029-2019. Microbiol Spectr. 2019. PMID: 31298207 Free PMC article. Review.
Cited by
-
The current landscape of microRNAs (miRNAs) in bacterial pneumonia: opportunities and challenges.Cell Mol Biol Lett. 2022 Aug 19;27(1):70. doi: 10.1186/s11658-022-00368-y. Cell Mol Biol Lett. 2022. PMID: 35986232 Free PMC article. Review.
-
Functional Analysis of EspM, an ESX-1-Associated Transcription Factor in Mycobacterium marinum.J Bacteriol. 2022 Dec 20;204(12):e0023322. doi: 10.1128/jb.00233-22. Epub 2022 Nov 30. J Bacteriol. 2022. PMID: 36448785 Free PMC article.
-
From accurate genome sequence to biotechnological application: The thermophile Mycolicibacterium hassiacum as experimental model.Microb Biotechnol. 2024 Jan;17(1):e14290. doi: 10.1111/1751-7915.14290. Epub 2023 Jul 27. Microb Biotechnol. 2024. PMID: 37498289 Free PMC article.
-
A Small Protein but with Diverse Roles: A Review of EsxA in Mycobacterium-Host Interaction.Cells. 2021 Jun 30;10(7):1645. doi: 10.3390/cells10071645. Cells. 2021. PMID: 34209120 Free PMC article. Review.
-
Bacterial secretion systems: Networks of pathogenic regulation and adaptation in mycobacteria and beyond.PLoS Pathog. 2022 Jul 14;18(7):e1010610. doi: 10.1371/journal.ppat.1010610. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35834482 Free PMC article. No abstract available.
References
-
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR Jr.. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A 100:12420–12425. doi:10.1073/pnas.1635213100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

