NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells
- PMID: 32899774
- PMCID: PMC7565505
- DOI: 10.3390/cells9092032
NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells
Abstract
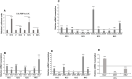
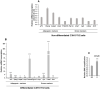
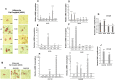
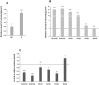
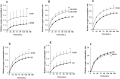
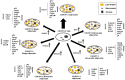
The NOTCH family of receptors and ligands is involved in numerous cell differentiation processes, including adipogenesis. We recently showed that overexpression of each of the four NOTCH receptors in 3T3-L1 preadipocytes enhances adipogenesis and modulates the acquisition of the mature adipocyte phenotype. We also revealed that DLK proteins modulate the adipogenesis of 3T3-L1 preadipocytes and mesenchymal C3H10T1/2 cells in an opposite way, despite their function as non-canonical inhibitory ligands of NOTCH receptors. In this work, we used multipotent C3H10T1/2 cells as an adipogenic model. We used standard adipogenic procedures and analyzed different parameters by using quantitative-polymerase chain reaction (qPCR), quantitative reverse transcription-polymerase chain reaction (qRT-PCR), luciferase, Western blot, and metabolic assays. We revealed that C3H10T1/2 multipotent cells show higher levels of NOTCH receptors expression and activity and lower Dlk gene expression levels than 3T3-L1 preadipocytes. We found that the overexpression of NOTCH receptors enhanced C3H10T1/2 adipogenesis levels, and the overexpression of NOTCH receptors and DLK (DELTA-like homolog) proteins modulated the conversion of cells towards a brown-like adipocyte phenotype. These and our prior results with 3T3-L1 preadipocytes strengthen the idea that, depending on the cellular context, a precise and highly regulated level of global NOTCH signaling is necessary to allow adipogenesis and determine the mature adipocyte phenotype.
Keywords: EGF-like proteins; adipogenic differentiation; brown-like adipocytes; mesenchymal cells; preadipocytes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
DLK proteins modulate NOTCH signaling to influence a brown or white 3T3-L1 adipocyte fate.Sci Rep. 2018 Nov 16;8(1):16923. doi: 10.1038/s41598-018-35252-3. Sci Rep. 2018. PMID: 30446682 Free PMC article.
-
DLK1 and DLK2, two non-canonical ligands of NOTCH receptors, differentially modulate the osteogenic differentiation of mesenchymal C3H10T1/2 cells.Biol Res. 2024 Oct 30;57(1):77. doi: 10.1186/s40659-024-00561-7. Biol Res. 2024. PMID: 39473022 Free PMC article.
-
The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other's activities.Biochim Biophys Acta. 2011 Jun;1813(6):1153-64. doi: 10.1016/j.bbamcr.2011.03.004. Epub 2011 Mar 15. Biochim Biophys Acta. 2011. PMID: 21419176
-
The Role of Pref-1 during Adipogenic Differentiation: An Overview of Suggested Mechanisms.Int J Mol Sci. 2020 Jun 9;21(11):4104. doi: 10.3390/ijms21114104. Int J Mol Sci. 2020. PMID: 32526833 Free PMC article. Review.
-
Roles of Notch Signaling in Adipocyte Progenitor Cells and Mature Adipocytes.J Cell Physiol. 2017 Jun;232(6):1258-1261. doi: 10.1002/jcp.25697. Epub 2017 Jan 5. J Cell Physiol. 2017. PMID: 27869309 Review.
Cited by
-
Gene expression and characterization of clonally derived murine embryonic brown and brite adipocytes.FEBS Open Bio. 2024 Sep;14(9):1503-1525. doi: 10.1002/2211-5463.13861. Epub 2024 Jul 7. FEBS Open Bio. 2024. PMID: 38972757 Free PMC article.
-
Different Expression Levels of DLK2 Inhibit NOTCH Signaling and Inversely Modulate MDA-MB-231 Breast Cancer Tumor Growth In Vivo.Int J Mol Sci. 2022 Jan 29;23(3):1554. doi: 10.3390/ijms23031554. Int J Mol Sci. 2022. PMID: 35163478 Free PMC article.
-
Notch pathway inhibitor DAPT accelerates in vitro proliferation and adipogenesis in infantile hemangioma stem cells.Oncol Lett. 2021 Dec;22(6):854. doi: 10.3892/ol.2021.13115. Epub 2021 Oct 26. Oncol Lett. 2021. PMID: 34777588 Free PMC article.
-
The effects of β-catenin on cardiomyogenesis via Islet-1 and MLIP ubiquitination.Exp Biol Med (Maywood). 2022 Nov;247(21):1956-1967. doi: 10.1177/15353702221119792. Epub 2022 Sep 12. Exp Biol Med (Maywood). 2022. PMID: 36112854 Free PMC article.
-
Emerging Roles of DLK1 in the Stem Cell Niche and Cancer Stemness.J Histochem Cytochem. 2022 Jan;70(1):17-28. doi: 10.1369/00221554211048951. Epub 2021 Oct 4. J Histochem Cytochem. 2022. PMID: 34606325 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

