The Epilepsy-Related Protein PCDH19 Regulates Tonic Inhibition, GABAAR Kinetics, and the Intrinsic Excitability of Hippocampal Neurons
- PMID: 32880860
- PMCID: PMC7541378
- DOI: 10.1007/s12035-020-02099-7
The Epilepsy-Related Protein PCDH19 Regulates Tonic Inhibition, GABAAR Kinetics, and the Intrinsic Excitability of Hippocampal Neurons
Abstract
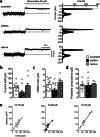
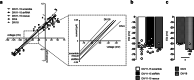
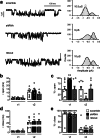
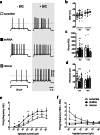
PCDH19 encodes for protocadherin-19 (PCDH19), a cell-adhesion molecule of the cadherin superfamily preferentially expressed in the brain. PCDH19 mutations cause a neurodevelopmental syndrome named epileptic encephalopathy, early infantile, 9 (EIEE9) characterized by seizures associated with cognitive and behavioral deficits. We recently reported that PCDH19 binds the alpha subunits of GABAA receptors (GABAARs), modulating their surface availability and miniature inhibitory postsynaptic currents (mIPSCs). Here, we investigated whether PCDH19 regulatory function on GABAARs extends to the extrasynaptic receptor pool that mediates tonic current. In fact, the latter shapes neuronal excitability and network properties at the base of information processing. By combining patch-clamp recordings in whole-cell and cell-attached configurations, we provided a functional characterization of primary hippocampal neurons from embryonic rats of either sex expressing a specific PCDH19 short hairpin (sh)RNA. We first demonstrated that PCDH19 downregulation reduces GABAAR-mediated tonic current, evaluated by current shift and baseline noise analysis. Next, by single-channel recordings, we showed that PCDH19 regulates GABAARs kinetics without altering their conductance. In particular, GABAARs of shRNA-expressing neurons preferentially exhibit brief openings at the expense of long ones, thus displaying a flickering behavior. Finally, we showed that PCDH19 downregulation reduces the rheobase and increases the frequency of action potential firing, thus indicating neuronal hyperexcitability. These findings establish PCDH19 as a critical determinant of GABAAR-mediated tonic transmission and GABAARs gating, and provide the first mechanistic insights into PCDH19-related hyperexcitability and comorbidities.
Keywords: ASD; GABAAR; Hyperexcitability; Intellectual disability; PCDH19.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
The female epilepsy protein PCDH19 is a new GABAAR-binding partner that regulates GABAergic transmission as well as migration and morphological maturation of hippocampal neurons.Hum Mol Genet. 2018 Mar 15;27(6):1027-1038. doi: 10.1093/hmg/ddy019. Hum Mol Genet. 2018. PMID: 29360992 Free PMC article.
-
Enhanced GABAA-Mediated Tonic Inhibition in Auditory Thalamus of Rats with Behavioral Evidence of Tinnitus.J Neurosci. 2015 Jun 24;35(25):9369-80. doi: 10.1523/JNEUROSCI.5054-14.2015. J Neurosci. 2015. PMID: 26109660 Free PMC article.
-
Differential Coassembly of α1-GABAARs Associated with Epileptic Encephalopathy.J Neurosci. 2020 Jul 15;40(29):5518-5530. doi: 10.1523/JNEUROSCI.2748-19.2020. Epub 2020 Jun 8. J Neurosci. 2020. PMID: 32513829 Free PMC article.
-
The Role of Protocadherin 19 (PCDH19) in Neurodevelopment and in the Pathophysiology of Early Infantile Epileptic Encephalopathy-9 (EIEE9).Dev Neurobiol. 2019 Jan;79(1):75-84. doi: 10.1002/dneu.22654. Epub 2019 Jan 18. Dev Neurobiol. 2019. PMID: 30431232 Review.
-
Extrasynaptic δ-subunit containing GABAA receptors.J Integr Neurosci. 2021 Mar 30;20(1):173-184. doi: 10.31083/j.jin.2021.01.284. J Integr Neurosci. 2021. PMID: 33834705 Review.
Cited by
-
PCDH19-related epilepsy in mosaic males: The phenotypic implication of genotype and variant allele frequency.Front Neurol. 2022 Nov 3;13:1041509. doi: 10.3389/fneur.2022.1041509. eCollection 2022. Front Neurol. 2022. PMID: 36408521 Free PMC article.
-
Perturbation of Cortical Excitability in a Conditional Model of PCDH19 Disorder.Cells. 2022 Jun 16;11(12):1939. doi: 10.3390/cells11121939. Cells. 2022. PMID: 35741068 Free PMC article.
-
Dissecting the Role of PCDH19 in Clustering Epilepsy by Exploiting Patient-Specific Models of Neurogenesis.J Clin Med. 2021 Jun 23;10(13):2754. doi: 10.3390/jcm10132754. J Clin Med. 2021. PMID: 34201522 Free PMC article.
-
Multiomic analysis implicates nuclear hormone receptor signalling in clustering epilepsy.Transl Psychiatry. 2024 Jan 27;14(1):65. doi: 10.1038/s41398-024-02783-5. Transl Psychiatry. 2024. PMID: 38280856 Free PMC article. Review.
-
A Case Report of Parental Germline Mosaicism in the PCDH19 Gene of Two Iranian Siblings.Basic Clin Neurosci. 2024 Jul-Aug;15(4):541-552. doi: 10.32598/bcn.2023.5507.1. Epub 2024 Jul 1. Basic Clin Neurosci. 2024. PMID: 39553263 Free PMC article.
References
-
- Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, Bomar J, Sutton E, Vandeleur L, Shoubridge C, Edkins S, Turner SJ, Stevens C, O'Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Jones D, Lee R, Madison M, Mironenko T, Varian J, West S, Widaa S, Wray P, Teague J, Dicks E, Butler A, Menzies A, Jenkinson A, Shepherd R, Gusella JF, Afawi Z, Mazarib A, Neufeld MY, Kivity S, Lev D, Lerman-Sagie T, Korczyn AD, Derry CP, Sutherland GR, Friend K, Shaw M, Corbett M, Kim HG, Geschwind DH, Thomas P, Haan E, Ryan S, McKee S, Berkovic SF, Futreal PA, Stratton MR, Mulley JC, Gécz J. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. - DOI - PMC - PubMed
-
- Kolc KL, Sadleir LG, Scheffer IE, Ivancevic A, Roberts R, Pham DH, Gecz J. A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol Psychiatry. 2018;24:241–251. doi: 10.1038/s41380-018-0066-9. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

