Structural alphabets for conformational analysis of nucleic acids available at dnatco.datmos.org
- PMID: 32876056
- PMCID: PMC7466747
- DOI: 10.1107/S2059798320009389
Structural alphabets for conformational analysis of nucleic acids available at dnatco.datmos.org
Abstract
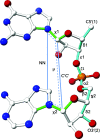
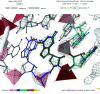
A detailed description of the dnatco.datmos.org web server implementing the universal structural alphabet of nucleic acids is presented. It is capable of processing any mmCIF- or PDB-formatted files containing DNA or RNA molecules; these can either be uploaded by the user or supplied as the wwPDB or PDB-REDO structural database access code. The web server performs an assignment of the nucleic acid conformations and presents the results for the intuitive annotation, validation, modeling and refinement of nucleic acids.
Keywords: annotation; nucleic acids; refinement; structural alphabets; validation.
open access.
Figures





Similar articles
-
DNATCO: assignment of DNA conformers at dnatco.org.Nucleic Acids Res. 2016 Jul 8;44(W1):W284-7. doi: 10.1093/nar/gkw381. Epub 2016 May 5. Nucleic Acids Res. 2016. PMID: 27150812 Free PMC article.
-
A unified dinucleotide alphabet describing both RNA and DNA structures.Nucleic Acids Res. 2020 Jun 19;48(11):6367-6381. doi: 10.1093/nar/gkaa383. Nucleic Acids Res. 2020. PMID: 32406923 Free PMC article.
-
WebSTAR3D: a web server for RNA 3D structural alignment.Bioinformatics. 2016 Dec 1;32(23):3673-3675. doi: 10.1093/bioinformatics/btw502. Epub 2016 Aug 6. Bioinformatics. 2016. PMID: 27497443 Free PMC article.
-
PDBx/mmCIF Ecosystem: Foundational Semantic Tools for Structural Biology.J Mol Biol. 2022 Jun 15;434(11):167599. doi: 10.1016/j.jmb.2022.167599. Epub 2022 Apr 20. J Mol Biol. 2022. PMID: 35460671 Free PMC article. Review.
-
Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive.Methods Mol Biol. 2017;1607:627-641. doi: 10.1007/978-1-4939-7000-1_26. Methods Mol Biol. 2017. PMID: 28573592 Free PMC article. Review.
Cited by
-
Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.Acta Crystallogr D Struct Biol. 2020 Dec 1;76(Pt 12):1233-1243. doi: 10.1107/S2059798320014151. Epub 2020 Nov 24. Acta Crystallogr D Struct Biol. 2020. PMID: 33263329 Free PMC article.
-
Knowledge-based prediction of DNA hydration using hydrated dinucleotides as building blocks.Acta Crystallogr D Struct Biol. 2022 Aug 1;78(Pt 8):1032-1045. doi: 10.1107/S2059798322006234. Epub 2022 Jul 21. Acta Crystallogr D Struct Biol. 2022. PMID: 35916227 Free PMC article.
-
FURNA: A database for functional annotations of RNA structures.PLoS Biol. 2024 Jul 29;22(7):e3002476. doi: 10.1371/journal.pbio.3002476. eCollection 2024 Jul. PLoS Biol. 2024. PMID: 39074139 Free PMC article.
-
Are kuravirus capsid diameters quantized? The first all-atom genome tracing method for double-stranded DNA viruses.Nucleic Acids Res. 2024 Feb 9;52(3):e12. doi: 10.1093/nar/gkad1153. Nucleic Acids Res. 2024. PMID: 38084886 Free PMC article.
-
The Nucleic Acid Knowledgebase: a new portal for 3D structural information about nucleic acids.Nucleic Acids Res. 2024 Jan 5;52(D1):D245-D254. doi: 10.1093/nar/gkad957. Nucleic Acids Res. 2024. PMID: 37953312 Free PMC article.
References
-
- Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J. D. & Zardecki, C. (2002). Acta Cryst. D58, 899–907. - PubMed
-
- Berman, H. M., Westbrook, J., Feng, Z., Iype, L., Schneider, B. & Zardecki, C. (2002). Acta Cryst. D58, 889–898. - PubMed
-
- Biedermannová, L. & Schneider, B. (2016). Biochim. Biophys. Acta, 1860, 1821–1835. - PubMed
-
- Burley, S. K., Berman, H. M., Bhikadiya, C., Bi, C., Chen, L., Di Costanzo, L., Christie, C., Dalenberg, K., Duarte, J. M., Dutta, S., Feng, Z., Ghosh, S., Goodsell, D. S., Green, R. K., Guranović, V., Guzenko, D., Hudson, B. P., Kalro, T., Liang, Y., Lowe, R., Namkoong, H., Peisach, E., Periskova, I., Prlić, A., Randle, C., Rose, A., Rose, P., Sala, R., Sekharan, M., Shao, C., Tan, L., Tao, Y.-P., Valasatava, Y., Voigt, M., Westbrook, J., Woo, J., Yang, H., Young, J., Zhuravleva, M. & Zardecki, C. (2019). Nucleic Acids Res. 47, D464–D474. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- CZ.02.1.01/0.0/0.0/16_013/0001777/Ministerstvo Školství, Mládeže a Tělovýchovy
- CZ.1.05/1.1.00/02.0109/Ministerstvo Školství, Mládeže a Tělovýchovy
- LM2018131/Ministerstvo Školství, Mládeže a Tělovýchovy
- LTAUSA18197/Ministerstvo Školství, Mládeže a Tělovýchovy
- RVO 86652036/Akademie Věd České Republiky, Biotechnologický ústav, Akademie Věd České Republiky
LinkOut - more resources
Full Text Sources

