Thyroid Hormone Induces DNA Demethylation in Xenopus Tadpole Brain
- PMID: 32865566
- PMCID: PMC7947600
- DOI: 10.1210/endocr/bqaa155
Thyroid Hormone Induces DNA Demethylation in Xenopus Tadpole Brain
Abstract
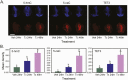
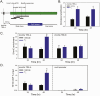
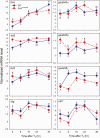
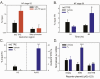
Thyroid hormone (T3) plays pivotal roles in vertebrate development, acting via nuclear T3 receptors (TRs) that regulate gene transcription by promoting post-translational modifications to histones. Methylation of cytosine residues in deoxyribonucleic acid (DNA) also modulates gene transcription, and our recent finding of predominant DNA demethylation in the brain of Xenopus tadpoles at metamorphosis, a T3-dependent developmental process, caused us to hypothesize that T3 induces these changes in vivo. Treatment of premetamorphic tadpoles with T3 for 24 or 48 hours increased immunoreactivity in several brain regions for the DNA demethylation intermediates 5-hydroxymethylcytosine (5-hmC) and 5-carboxylcytosine, and the methylcytosine dioxygenase ten-eleven translocation 3 (TET3). Thyroid hormone treatment induced locus-specific DNA demethylation in proximity to known T3 response elements within the DNA methyltransferase 3a and Krüppel-like factor 9 genes, analyzed by 5-hmC immunoprecipitation and methylation sensitive restriction enzyme digest. Chromatin-immunoprecipitation (ChIP) assay showed that T3 induced TET3 recruitment to these loci. Furthermore, the messenger ribonucleic acid for several genes encoding DNA demethylation enzymes were induced by T3 in a time-dependent manner in tadpole brain. A TR ChIP-sequencing experiment identified putative TR binding sites at several of these genes, and we provide multiple lines of evidence to support that tet2 contains a bona fide T3 response element. Our findings show that T3 can promote DNA demethylation in developing tadpole brain, in part by promoting TET3 recruitment to discrete genomic regions, and by inducing genes that encode DNA demethylation enzymes.
Keywords: DNA methylation; Xenopus; brain development; chromatin; metamorphosis; thyroid hormone.
© Endocrine Society 2020. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Similar articles
-
DNA methylation dynamics underlie metamorphic gene regulation programs in Xenopus tadpole brain.Dev Biol. 2020 Jun 15;462(2):180-196. doi: 10.1016/j.ydbio.2020.03.013. Epub 2020 Mar 31. Dev Biol. 2020. PMID: 32240642 Free PMC article.
-
A Mechanism to Enhance Cellular Responsivity to Hormone Action: Krüppel-Like Factor 9 Promotes Thyroid Hormone Receptor-β Autoinduction During Postembryonic Brain Development.Endocrinology. 2016 Apr;157(4):1683-93. doi: 10.1210/en.2015-1980. Epub 2016 Feb 17. Endocrinology. 2016. PMID: 26886257 Free PMC article.
-
Thyroid hormone activates Xenopus MBD3 gene via an intronic TRE in vivo.Front Biosci (Landmark Ed). 2020 Jan 1;25(3):437-451. doi: 10.2741/4812. Front Biosci (Landmark Ed). 2020. PMID: 31585895 Free PMC article.
-
Autoinduction of nuclear receptor genes and its significance.J Steroid Biochem Mol Biol. 1993 Aug;46(2):105-19. doi: 10.1016/0960-0760(93)90286-6. J Steroid Biochem Mol Biol. 1993. PMID: 8664159 Review.
-
Dual functions of thyroid hormone receptors during Xenopus development.Comp Biochem Physiol B Biochem Mol Biol. 2000 Jun;126(2):199-211. doi: 10.1016/s0305-0491(00)00198-x. Comp Biochem Physiol B Biochem Mol Biol. 2000. PMID: 10874167 Review.
Cited by
-
TET3 Mediates 5hmC Level and Promotes Tumorigenesis by Activating AMPK Pathway in Papillary Thyroid Cancer.Int J Endocrinol. 2022 Jun 15;2022:2658727. doi: 10.1155/2022/2658727. eCollection 2022. Int J Endocrinol. 2022. PMID: 35755313 Free PMC article.
-
Cellular Iron Deficiency Disrupts Thyroid Hormone Regulated Gene Expression in Developing Hippocampal Neurons.J Nutr. 2024 Jan;154(1):49-59. doi: 10.1016/j.tjnut.2023.11.007. Epub 2023 Nov 19. J Nutr. 2024. PMID: 37984740 Free PMC article.
-
Involvement of Thyroid Hormones in Brain Development and Cancer.Cancers (Basel). 2021 May 30;13(11):2693. doi: 10.3390/cancers13112693. Cancers (Basel). 2021. PMID: 34070729 Free PMC article. Review.
-
Thyroid hormone links environmental signals to DNA methylation.Philos Trans R Soc Lond B Biol Sci. 2024 Mar 25;379(1898):20220506. doi: 10.1098/rstb.2022.0506. Epub 2024 Feb 5. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 38310936 Review.
-
Epigenetic developmental programming and intergenerational effects of thyroid hormones.Vitam Horm. 2023;122:23-49. doi: 10.1016/bs.vh.2023.01.003. Epub 2023 Feb 9. Vitam Horm. 2023. PMID: 36863795 Free PMC article. Review.
References
-
- Bernal J; Thyroid Hormones and Brain Development . Hormones, Brain and Behavior, Vol 5: Development of Hormone-Behavior Relationships. 3rd ed. Auger AP, Auger CJ, Pfaff DW, Joels M, eds. San Diego: Academic Press, Inc; 2017: 159-184.
-
- Porterfield SP, Hendrich CE. The role of thyroid hormones in prenatal and neonatal neurological development–current perspectives. Endocr Rev. 1993;14(1):94–106. - PubMed
-
- Shi Y-B Amphibian metamorphosis: from morphology to molecular biology. New York: Wiley-Liss; 2000.
-
- Vennstrom B, Liu H, Forrest D. Thyroid hormone receptors. In: Bunce CM, Campbell MJ, eds. Nuclear Receptors: Current Concepts and Future Challenges. New York, NY: Springer; 82010:183–201.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

