Antigenic properties of dense granule antigen 12 protein using bioinformatics tools in order to improve vaccine design against Toxoplasma gondii
- PMID: 32864364
- PMCID: PMC7445328
- DOI: 10.7774/cevr.2020.9.2.81
Antigenic properties of dense granule antigen 12 protein using bioinformatics tools in order to improve vaccine design against Toxoplasma gondii
Abstract
Purpose: Toxoplasma gondii is an opportunistic parasite infecting all warm-blooded animals including humans. The dense granule antigens (GRAs) play an important role in parasite survival and virulence and in forming the parasitophorous vacuole. Identification of protein characteristics increases our knowledge about them and leads to develop the vaccine and diagnostic studies.
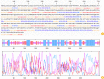
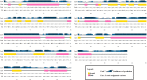
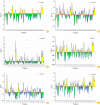
Materials and methods: This paper gave a comprehensive definition of the important aspects of GRA12 protein, including physico-chemical features, a transmembrane domain, subcellular position, secondary and tertiary structure, potential epitopes of B-cells and T-cells, and other important features of this protein using different and reliable bioinformatics methods to determine potential epitopes for designing of a high-efficient vaccine.
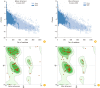
Results: The findings showed that GRA12 protein had 53 potential post-translational modification sites. Also, only one transmembrane domain was recognized for this protein. The secondary structure of GRA12 protein comprises 35.55% alpha-helix, 19.50% extended strand, and 44.95% random coil. Moreover, several potential B- and T-cell epitopes were identified for GRA12. Based on the results of the Ramachandran plot, 79.26% of amino acid residues were located in favored, 11.85% in allowed and 8.89% in outlier regions. Furthermore, the results of the antigenicity and allergenicity assessment noted that GRA12 is immunogenic and non-allergenic.
Conclusion: This research provided important basic and conceptual data on GRA12 to develop an effective vaccine against acute and chronic toxoplasmosis for further in vivo investigations. More studies are required on vaccine development using the GRA12 alone or combined with other antigens in the future.
Keywords: Computational biology; Epitope mapping; Toxoplasma gondii; Vaccines.
© Korean Vaccine Society.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures




Similar articles
-
Bioinformatics analysis of ROP8 protein to improve vaccine design against Toxoplasma gondii.Infect Genet Evol. 2018 Aug;62:193-204. doi: 10.1016/j.meegid.2018.04.033. Epub 2018 Apr 26. Infect Genet Evol. 2018. PMID: 29705360
-
Structural predication and antigenic analysis of ROP16 protein utilizing immunoinformatics methods in order to identification of a vaccine against Toxoplasma gondii: An in silico approach.Microb Pathog. 2020 Feb 19;142:104079. doi: 10.1016/j.micpath.2020.104079. Online ahead of print. Microb Pathog. 2020. PMID: 32084578
-
In silico analysis and structural vaccinology prediction of Toxoplasma gondii ROP41 gene via immunoinformatics methods as a vaccine candidate.Curr Res Transl Med. 2024 Oct 16;73(1):103475. doi: 10.1016/j.retram.2024.103475. Online ahead of print. Curr Res Transl Med. 2024. PMID: 39461097
-
Candidate antigenic epitopes for vaccination and diagnosis strategies of Toxoplasma gondii infection: A review.Microb Pathog. 2019 Dec;137:103788. doi: 10.1016/j.micpath.2019.103788. Epub 2019 Oct 9. Microb Pathog. 2019. PMID: 31605758 Review.
-
Rhoptry antigens as Toxoplasma gondii vaccine target.Clin Exp Vaccine Res. 2019 Jan;8(1):4-26. doi: 10.7774/cevr.2019.8.1.4. Epub 2019 Jan 31. Clin Exp Vaccine Res. 2019. PMID: 30775347 Free PMC article. Review.
Cited by
-
Immunoinformatic Analysis of Calcium-Dependent Protein Kinase 7 (CDPK7) Showed Potential Targets for Toxoplasma gondii Vaccine.J Parasitol Res. 2021 Jul 8;2021:9974509. doi: 10.1155/2021/9974509. eCollection 2021. J Parasitol Res. 2021. PMID: 34336254 Free PMC article.
-
More Than Seventy Years of Research (1948-November 2021) on Toxoplasma gondii in Iran: A Narrative Review.Iran J Parasitol. 2022 Apr-Jun;17(2):124-137. doi: 10.18502/ijpa.v17i2.9528. Iran J Parasitol. 2022. PMID: 36032751 Free PMC article. Review.
-
Protective efficacy of Toxoplasma gondii GRA12 or GRA7 recombinant proteins encapsulated in PLGA nanoparticles against acute Toxoplasma gondii infection in mice.Front Cell Infect Microbiol. 2023 Jul 12;13:1209755. doi: 10.3389/fcimb.2023.1209755. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37502604 Free PMC article.
-
Letter to the editor of Heliyon re: Bioinformatics-based prediction and screening of immunogenic epitopes of Toxoplasma gondii rhoptry proteins 7, 21 and 22 as candidate vaccine target.Heliyon. 2024 Jun 4;10(14):e32221. doi: 10.1016/j.heliyon.2024.e32221. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39113981 Free PMC article. No abstract available.
-
Immunoinformatics studies and design of a novel multi-epitope peptide vaccine against Toxoplasma gondii based on calcium-dependent protein kinases antigens through an in-silico analysis.Clin Exp Vaccine Res. 2024 Apr;13(2):146-154. doi: 10.7774/cevr.2024.13.2.146. Epub 2024 Apr 30. Clin Exp Vaccine Res. 2024. PMID: 38752002 Free PMC article.
References
-
- Shaddel M, Mirzaii Dizgah I, Sharif F. The prevalence of toxoplasmosis in Imam Reza Hospital blood bank samples, Tehran, Iran. Transfus Apher Sci. 2014;51:181–183. - PubMed
-
- Ghaffari AD, Dalimi A. Molecular identification of Toxoplasma gondii in the native slaughtered cattle of Tehran province, Iran. J Food Qual Hazards Control. 2019;6:153–161.
-
- Foroutan M, Ghaffarifar F, Sharifi Z, Dalimi A, Pirestani M. Bioinformatics analysis of ROP8 protein to improve vaccine design against Toxoplasma gondii. Infect Genet Evol. 2018;62:193–204. - PubMed
-
- Chaichan P, Mercier A, Galal L, et al. Geographical distribution of Toxoplasma gondii genotypes in Asia: a link with neighboring continents. Infect Genet Evol. 2017;53:227–238. - PubMed
LinkOut - more resources
Full Text Sources

