Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium
- PMID: 32858958
- PMCID: PMC7565861
- DOI: 10.3390/genes11091002
Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium
Abstract
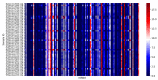
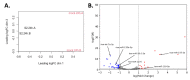
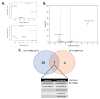
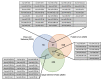
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), an RNA virus, is responsible for the coronavirus disease 2019 (COVID-19) pandemic of 2020. Experimental evidence suggests that microRNA can mediate an intracellular defence mechanism against some RNA viruses. The purpose of this study was to identify microRNA with predicted binding sites in the SARS-CoV-2 genome, compare these to their microRNA expression profiles in lung epithelial tissue and make inference towards possible roles for microRNA in mitigating coronavirus infection. We hypothesize that high expression of specific coronavirus-targeting microRNA in lung epithelia may protect against infection and viral propagation, conversely, low expression may confer susceptibility to infection. We have identified 128 human microRNA with potential to target the SARS-CoV-2 genome, most of which have very low expression in lung epithelia. Six of these 128 microRNA are differentially expressed upon in vitro infection of SARS-CoV-2. Additionally, 28 microRNA also target the SARS-CoV genome while 23 microRNA target the MERS-CoV genome. We also found that a number of microRNA are commonly identified in two other studies. Further research into identifying bona fide coronavirus targeting microRNA will be useful in understanding the importance of microRNA as a cellular defence mechanism against pathogenic coronavirus infections.
Keywords: SARS-CoV-2; cellular antiviral defence; coronavirus; lung epithelia; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Conserved Genomic Terminals of SARS-CoV-2 as Coevolving Functional Elements and Potential Therapeutic Targets.mSphere. 2020 Nov 25;5(6):e00754-20. doi: 10.1128/mSphere.00754-20. mSphere. 2020. PMID: 33239366 Free PMC article.
-
The Prediction of miRNAs in SARS-CoV-2 Genomes: hsa-miR Databases Identify 7 Key miRs Linked to Host Responses and Virus Pathogenicity-Related KEGG Pathways Significant for Comorbidities.Viruses. 2020 Jun 4;12(6):614. doi: 10.3390/v12060614. Viruses. 2020. PMID: 32512929 Free PMC article.
-
Gene signatures and potential therapeutic targets of Middle East respiratory syndrome coronavirus (MERS-CoV)-infected human lung adenocarcinoma epithelial cells.J Microbiol Immunol Infect. 2021 Oct;54(5):845-857. doi: 10.1016/j.jmii.2021.03.007. Epub 2021 Mar 26. J Microbiol Immunol Infect. 2021. PMID: 34176764 Free PMC article.
-
Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach.Biochim Biophys Acta Mol Basis Dis. 2020 Oct 1;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878. Epub 2020 Jun 13. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 32544429 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
Cited by
-
The relationship between microRNAs and COVID-19 complications.Noncoding RNA Res. 2024 Aug 22;10:16-24. doi: 10.1016/j.ncrna.2024.08.007. eCollection 2025 Feb. Noncoding RNA Res. 2024. PMID: 39296641 Free PMC article. Review.
-
Nucleic Acid-Based COVID-19 Therapy Targeting Cytokine Storms: Strategies to Quell the Storm.J Pers Med. 2022 Mar 3;12(3):386. doi: 10.3390/jpm12030386. J Pers Med. 2022. PMID: 35330388 Free PMC article. Review.
-
microRNAs and Inflammatory Immune Response in SARS-CoV-2 Infection: A Narrative Review.Life (Basel). 2022 Feb 15;12(2):288. doi: 10.3390/life12020288. Life (Basel). 2022. PMID: 35207576 Free PMC article. Review.
-
Identifying Putative Causal Links between MicroRNAs and Severe COVID-19 Using Mendelian Randomization.Cells. 2021 Dec 11;10(12):3504. doi: 10.3390/cells10123504. Cells. 2021. PMID: 34944012 Free PMC article.
-
Increased Serum Mir-150-3p Expression Is Associated with Radiological Lung Injury Improvement in Patients with COVID-19.Viruses. 2022 Jun 23;14(7):1363. doi: 10.3390/v14071363. Viruses. 2022. PMID: 35891345 Free PMC article.
References
-
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

