Characterization of BRCA1/2-Directed ceRNA Network Identifies a Novel Three-lncRNA Signature to Predict Prognosis and Chemo-Response in Ovarian Cancer Patients With Wild-Type BRCA1/2
- PMID: 32850807
- PMCID: PMC7403448
- DOI: 10.3389/fcell.2020.00680
Characterization of BRCA1/2-Directed ceRNA Network Identifies a Novel Three-lncRNA Signature to Predict Prognosis and Chemo-Response in Ovarian Cancer Patients With Wild-Type BRCA1/2
Abstract
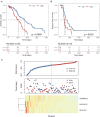
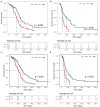
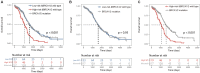
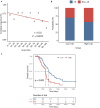
Long non-coding RNAs (lncRNAs) have been reported to interact with BRCA1/2 to regulate homologous recombination (HR) by diverse mechanisms in ovarian cancers (OvCa). However, genome-wide screening of BRCA1/2-related lncRNAs and their clinical significance is still unexplored. In this study, we constructed a global BRCA1/2-directed lncRNA-associated ceRNA network by integrating paired lncRNA expression profiles, miRNA expression profiles, and BRCA1/2 expression profiles in BRCA1/2 wild-type patients and identified 111 BRCA1/2-related lncRNAs. Using the stepwise regression and Cox regression analysis, we developed a BRCA1/2-directed lncRNA signature (BRCALncSig), composing of three lncRNAs (LINC01619, DLX6-AS1, and AC004943.2) from the list of 111 BRCA1/2-related lncRNAs, which was an independent prognostic factor and was able to classify the patients into high- and low-risk groups with significantly different survival in the training dataset (HR = 2.73, 95 CI 1.65-4.51, p < 0.001). The prognostic performance of the BRCALncSig was further validated in the testing dataset (HR = 1.9, 95 CI 1.21-2.99, p = 0.005) and entire TCGA dataset (HR = 2.17, 95 CI 1.56-3.01, p < 0.001). Furthermore, the BRCALncSig is associated with chemo-response and was also capable of discriminating nonequivalent outcomes for patients achieving complete response (CR) (log-rank p = 0.003). Functional analyses suggested that mRNAs co-expressed with the BRCALncSig were enriched in cancer-related or cell proliferation-related biological processes and pathways. In summary, our study highlighted the clinical implication of BRCA1/2-directed lncRNAs in the prognosis and treatment response of BRCA1/2 wild-type patients.
Keywords: BRCA1/2; ceRNA; long non-coding RNAs; ovarian cancers; signature.
Copyright © 2020 Zhang, Wang, Zhu and Wu.
Figures

Similar articles
-
Expression profiles analysis reveals an integrated miRNA-lncRNA signature to predict survival in ovarian cancer patients with wild-type BRCA1/2.Oncotarget. 2017 Jul 26;8(40):68483-68492. doi: 10.18632/oncotarget.19590. eCollection 2017 Sep 15. Oncotarget. 2017. PMID: 28978132 Free PMC article.
-
Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer.Oncotarget. 2016 May 31;7(22):32433-48. doi: 10.18632/oncotarget.8653. Oncotarget. 2016. PMID: 27074572 Free PMC article.
-
Systematic analysis of lncRNA-miRNA-mRNA competing endogenous RNA network identifies four-lncRNA signature as a prognostic biomarker for breast cancer.J Transl Med. 2018 Sep 27;16(1):264. doi: 10.1186/s12967-018-1640-2. J Transl Med. 2018. PMID: 30261893 Free PMC article.
-
Systematic analysis reveals a lncRNA-mRNA co-expression network associated with platinum resistance in high-grade serous ovarian cancer.Invest New Drugs. 2018 Apr;36(2):187-194. doi: 10.1007/s10637-017-0523-3. Epub 2017 Oct 30. Invest New Drugs. 2018. PMID: 29082457
-
Analysis of lncRNA-Associated ceRNA Network Reveals Potential lncRNA Biomarkers in Human Colon Adenocarcinoma.Cell Physiol Biochem. 2018;49(5):1778-1791. doi: 10.1159/000493623. Epub 2018 Sep 19. Cell Physiol Biochem. 2018. PMID: 30231249
Cited by
-
Identification of lncRNAs involved in response to ionizing radiation in fibroblasts of long-term survivors of childhood cancer and cancer-free controls.Front Oncol. 2023 Apr 27;13:1158176. doi: 10.3389/fonc.2023.1158176. eCollection 2023. Front Oncol. 2023. PMID: 37182169 Free PMC article.
-
Development and Verification of an Autophagy-Related lncRNA Signature to Predict Clinical Outcomes and Therapeutic Responses in Ovarian Cancer.Front Med (Lausanne). 2021 Oct 4;8:715250. doi: 10.3389/fmed.2021.715250. eCollection 2021. Front Med (Lausanne). 2021. PMID: 34671615 Free PMC article.
-
Long Non-coding RNA LINC01969 Promotes Ovarian Cancer by Regulating the miR-144-5p/LARP1 Axis as a Competing Endogenous RNA.Front Cell Dev Biol. 2021 Feb 4;8:625730. doi: 10.3389/fcell.2020.625730. eCollection 2020. Front Cell Dev Biol. 2021. PMID: 33614632 Free PMC article.
References
-
- Bao S., Zhao H., Yuan J., Fan D., Zhang Z., Su J., et al. (2019). Computational identification of mutator-derived lncRNA signatures of genome instability for improving the clinical outcome of cancers: a case study in breast cancer. Brief. Bioinform. [Epub ahead of print]. 10.1093/bib/bbz118 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

