Measuring the Inducible, Replication-Competent HIV Reservoir Using an Ultra-Sensitive p24 Readout, the Digital ELISA Viral Outgrowth Assay
- PMID: 32849659
- PMCID: PMC7423995
- DOI: 10.3389/fimmu.2020.01971
Measuring the Inducible, Replication-Competent HIV Reservoir Using an Ultra-Sensitive p24 Readout, the Digital ELISA Viral Outgrowth Assay
Abstract
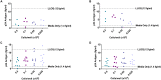
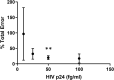
Quantifying the inducible HIV reservoir provides an estimate of the frequency of quiescent HIV-infected cells in humans as well as in animal models, and can help ascertain the efficacy of latency reversing agents (LRAs). The quantitative viral outgrowth assay (QVOA) is used to measure inducible, replication competent HIV and generate estimations of reservoir size. However, traditional QVOA is time and labor intensive and requires large amounts of lymphocytes. Given the importance of reproducible and accurate assessment of both reservoir size and LRA activity in cure strategies, efforts to streamline the QVOA are of high priority. We developed a modified QVOA, the Digital ELISA Viral Outgrowth or DEVO assay, with ultra-sensitive p24 readout, capable of femtogram detection of HIV p24 protein in contrast to the picogram limitations of traditional ELISA. For each DEVO assay, 8-12 × 106 resting CD4 + T cells from aviremic, ART-treated HIV + participants are plated in limiting dilution and maximally stimulated with PHA, IL-2 and uninfected allogeneic irradiated PBMC. CD8-depleted PHA blasts from an uninfected donor or HIV-permissive cells (e.g., Molt4/CCR5) are added to the cultures and virus allowed to amplify for 8-12 days. HIV p24 from culture supernatant is measured at day 8 by Simoa (single molecule array, ultra-sensitive p24 assay) confirmed at day 12, and infectious units per million CD4 + T cells (IUPM) are calculated using the maximum likelihood method. In all DEVO assays performed, HIV p24 was detected in the supernatant of cultures as early as 8 days post stimulation. Importantly, DEVO IUPM values at day 8 were comparable or higher than traditional QVOA IUPM values obtained at day 15. Interestingly, DEVO IUPM values were similar with or without the addition of allogeneic CD8-depleted target PHA blasts or HIV permissive cells traditionally used to expand virus. The DEVO assay uses fewer resting CD4 + T cells and provides an assessment of reservoir size in less time than standard QVOA. This assay offers a new platform to quantify replication competent HIV during limited cell availability. Other potential applications include evaluating LRA activity, and measuring clearance of infected cells during latency clearance assays.
Keywords: DEVO; HIV; IUPM; QVOA; outgrowth.
Copyright © 2020 Stuelke, James, Kirchherr, Allard, Baker, Kuruc, Gay, Margolis and Archin.
Figures
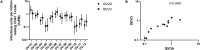



Similar articles
-
Ultrasensitive HIV-1 p24 Assay Detects Single Infected Cells and Differences in Reservoir Induction by Latency Reversal Agents.J Virol. 2017 Feb 28;91(6):e02296-16. doi: 10.1128/JVI.02296-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28077644 Free PMC article.
-
Improved assays to measure and characterize the inducible HIV reservoir.EBioMedicine. 2018 Oct;36:113-121. doi: 10.1016/j.ebiom.2018.09.036. Epub 2018 Oct 11. EBioMedicine. 2018. PMID: 30316868 Free PMC article.
-
Assessing the Suitability of Next-Generation Viral Outgrowth Assays to Measure Human Immunodeficiency Virus 1 Latent Reservoir Size.J Infect Dis. 2021 Oct 13;224(7):1209-1218. doi: 10.1093/infdis/jiaa089. J Infect Dis. 2021. PMID: 32147687 Free PMC article.
-
Low Inducibility of Latent Human Immunodeficiency Virus Type 1 Proviruses as a Major Barrier to Cure.J Infect Dis. 2021 Feb 15;223(12 Suppl 2):13-21. doi: 10.1093/infdis/jiaa649. J Infect Dis. 2021. PMID: 33586775 Free PMC article. Review.
-
Measuring integrated HIV DNA ex vivo and in vitro provides insights about how reservoirs are formed and maintained.Retrovirology. 2018 Feb 17;15(1):22. doi: 10.1186/s12977-018-0396-3. Retrovirology. 2018. PMID: 29452580 Free PMC article. Review.
Cited by
-
Nanoparticle delivery of Tat synergizes with classical latency reversal agents to express HIV antigen targets.Antimicrob Agents Chemother. 2024 Jul 9;68(7):e0020124. doi: 10.1128/aac.00201-24. Epub 2024 Jun 3. Antimicrob Agents Chemother. 2024. PMID: 38829049 Free PMC article.
-
Improved Detection of HIV Gag p24 Protein Using a Combined Immunoprecipitation and Digital ELISA Method.Front Microbiol. 2021 Mar 16;12:636703. doi: 10.3389/fmicb.2021.636703. eCollection 2021. Front Microbiol. 2021. PMID: 33796087 Free PMC article.
-
Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein.Talanta. 2022 Jan 1;236:122847. doi: 10.1016/j.talanta.2021.122847. Epub 2021 Sep 7. Talanta. 2022. PMID: 34635237 Free PMC article.
-
Application of ultrasensitive digital ELISA for p24 enables improved evaluation of HIV-1 reservoir diversity and growth kinetics in viral outgrowth assays.Sci Rep. 2023 Jul 6;13(1):10958. doi: 10.1038/s41598-023-37223-9. Sci Rep. 2023. PMID: 37414788 Free PMC article.
-
An ultrasensitive planar array p24 Gag ELISA to detect HIV-1 in diverse biological matrixes.Sci Rep. 2021 Dec 8;11(1):23682. doi: 10.1038/s41598-021-03072-7. Sci Rep. 2021. PMID: 34880361 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

