Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD
- PMID: 32848248
- PMCID: PMC8054479
- DOI: 10.1038/s41586-020-2633-x
Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD
Abstract
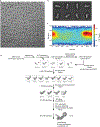
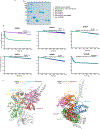
Mutation of C9orf72 is the most prevalent defect associated with amyotrophic lateral sclerosis and frontotemporal degeneration1. Together with hexanucleotide-repeat expansion2,3, haploinsufficiency of C9orf72 contributes to neuronal dysfunction4-6. Here we determine the structure of the C9orf72-SMCR8-WDR41 complex by cryo-electron microscopy. C9orf72 and SMCR8 both contain longin and DENN (differentially expressed in normal and neoplastic cells) domains7, and WDR41 is a β-propeller protein that binds to SMCR8 such that the whole structure resembles an eye slip hook. Contacts between WDR41 and the DENN domain of SMCR8 drive the lysosomal localization of the complex in conditions of amino acid starvation. The structure suggested that C9orf72-SMCR8 is a GTPase-activating protein (GAP), and we found that C9orf72-SMCR8-WDR41 acts as a GAP for the ARF family of small GTPases. These data shed light on the function of C9orf72 in normal physiology, and in amyotrophic lateral sclerosis and frontotemporal degeneration.
Figures




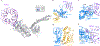



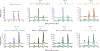



Similar articles
-
Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.Proc Natl Acad Sci U S A. 2020 May 5;117(18):9876-9883. doi: 10.1073/pnas.2002110117. Epub 2020 Apr 17. Proc Natl Acad Sci U S A. 2020. PMID: 32303654 Free PMC article.
-
The C9orf72-SMCR8-WDR41 complex is a GAP for small GTPases.Autophagy. 2020 Aug;16(8):1542-1543. doi: 10.1080/15548627.2020.1779473. Epub 2020 Jun 17. Autophagy. 2020. PMID: 32521185 Free PMC article.
-
Structural basis for the ARF GAP activity and specificity of the C9orf72 complex.Nat Commun. 2021 Jun 18;12(1):3786. doi: 10.1038/s41467-021-24081-0. Nat Commun. 2021. PMID: 34145292 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
The progress in C9orf72 research: ALS/FTD pathogenesis, functions and structure.Small GTPases. 2022 Jan;13(1):56-76. doi: 10.1080/21541248.2021.1892443. Epub 2021 Mar 5. Small GTPases. 2022. PMID: 33663328 Free PMC article. Review.
Cited by
-
Plug-and-socket mechanisms in nutrient sensing by lysosomal amino acid transporters.Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2102173118. doi: 10.1073/pnas.2102173118. Proc Natl Acad Sci U S A. 2021. PMID: 33723009 Free PMC article. No abstract available.
-
Receptor-like role for PQLC2 amino acid transporter in the lysosomal sensing of cationic amino acids.Proc Natl Acad Sci U S A. 2021 Feb 23;118(8):e2014941118. doi: 10.1073/pnas.2014941118. Proc Natl Acad Sci U S A. 2021. PMID: 33597295 Free PMC article.
-
The Interplay Between Autophagy and RNA Homeostasis: Implications for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia.Front Cell Dev Biol. 2022 Apr 28;10:838402. doi: 10.3389/fcell.2022.838402. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35573690 Free PMC article. Review.
-
Arginine-selective modulation of the lysosomal transporter PQLC2 through a gate-tuning mechanism.Proc Natl Acad Sci U S A. 2021 Aug 10;118(32):e2025315118. doi: 10.1073/pnas.2025315118. Proc Natl Acad Sci U S A. 2021. PMID: 34344826 Free PMC article.
-
Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.PLoS Biol. 2021 Jul 23;19(7):e3001344. doi: 10.1371/journal.pbio.3001344. eCollection 2021 Jul. PLoS Biol. 2021. PMID: 34297726 Free PMC article.
References
-
- Sivadasan R et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nature neuroscience 19, 1610–1618 (2016). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

