Structures of the Mononegavirales Polymerases
- PMID: 32847861
- PMCID: PMC7592205
- DOI: 10.1128/JVI.00175-20
Structures of the Mononegavirales Polymerases
Abstract
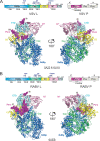
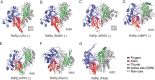
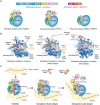
Mononegavirales, known as nonsegmented negative-sense (NNS) RNA viruses, are a class of pathogenic and sometimes deadly viruses that include rabies virus (RABV), human respiratory syncytial virus (HRSV), and Ebola virus (EBOV). Unfortunately, no effective vaccines and antiviral therapeutics against many Mononegavirales are currently available. Viral polymerases have been attractive and major antiviral therapeutic targets. Therefore, Mononegavirales polymerases have been extensively investigated for their structures and functions. Mononegavirales mimic RNA synthesis of their eukaryotic counterparts by utilizing multifunctional RNA polymerases to replicate entire viral genomes and transcribe viral mRNAs from individual viral genes as well as synthesize 5' methylated cap and 3' poly(A) tail of the transcribed viral mRNAs. The catalytic subunit large protein (L) and cofactor phosphoprotein (P) constitute the Mononegavirales polymerases. In this review, we discuss the shared and unique features of RNA synthesis, the monomeric multifunctional enzyme L, and the oligomeric multimodular adapter P of Mononegavirales We outline the structural analyses of the Mononegavirales polymerases since the first structure of the vesicular stomatitis virus (VSV) L protein determined in 2015 and highlight multiple high-resolution cryo-electron microscopy (cryo-EM) structures of the polymerases of Mononegavirales, namely, VSV, RABV, HRSV, human metapneumovirus (HMPV), and human parainfluenza virus (HPIV), that have been reported in recent months (2019 to 2020). We compare the structures of those polymerases grouped by virus family, illustrate the similarities and differences among those polymerases, and reveal the potential RNA synthesis mechanisms and models of highly conserved Mononegavirales We conclude by the discussion of remaining questions, evolutionary perspectives, and future directions.
Keywords: Mononegavirales polymerases; RNA-dependent RNA polymerase; cryo-EM structures; human metapneumovirus (HMPV); human respiratory syncytial virus (HRSV); rabies virus (RABV); vesicular stomatitis virus (VSV).
Copyright © 2020 Liang.
Figures






Similar articles
-
Structure of a rabies virus polymerase complex from electron cryo-microscopy.Proc Natl Acad Sci U S A. 2020 Jan 28;117(4):2099-2107. doi: 10.1073/pnas.1918809117. Epub 2020 Jan 17. Proc Natl Acad Sci U S A. 2020. PMID: 31953264 Free PMC article.
-
Oligomerization of the Vesicular Stomatitis Virus Phosphoprotein Is Dispensable for mRNA Synthesis but Facilitates RNA Replication.J Virol. 2020 Jun 16;94(13):e00115-20. doi: 10.1128/JVI.00115-20. Print 2020 Jun 16. J Virol. 2020. PMID: 32321813 Free PMC article.
-
Structure and function of negative-strand RNA virus polymerase complexes.Enzymes. 2021;50:21-78. doi: 10.1016/bs.enz.2021.09.002. Epub 2021 Nov 10. Enzymes. 2021. PMID: 34861938 Review.
-
Cryo-EM structure of the respiratory syncytial virus RNA polymerase.Nat Commun. 2020 Jan 17;11(1):368. doi: 10.1038/s41467-019-14246-3. Nat Commun. 2020. PMID: 31953395 Free PMC article.
-
New antiviral approaches for respiratory syncytial virus and other mononegaviruses: Inhibiting the RNA polymerase.Antiviral Res. 2016 Oct;134:63-76. doi: 10.1016/j.antiviral.2016.08.006. Epub 2016 Aug 27. Antiviral Res. 2016. PMID: 27575793 Review.
Cited by
-
Human Parainfluenza Virus 3 Phosphoprotein Is a Tetramer and Shares Structural and Interaction Features with Ebola Phosphoprotein VP35.Biomolecules. 2021 Oct 29;11(11):1603. doi: 10.3390/biom11111603. Biomolecules. 2021. PMID: 34827601 Free PMC article.
-
Optical Control of Mononegavirus Gene Expression and Replication.Methods Mol Biol. 2024;2808:35-56. doi: 10.1007/978-1-0716-3870-5_4. Methods Mol Biol. 2024. PMID: 38743361
-
The Nucleocapsid of Paramyxoviruses: Structure and Function of an Encapsidated Template.Viruses. 2021 Dec 9;13(12):2465. doi: 10.3390/v13122465. Viruses. 2021. PMID: 34960734 Free PMC article. Review.
-
CryoEM of Viral Ribonucleoproteins and Nucleocapsids of Single-Stranded RNA Viruses.Viruses. 2023 Feb 28;15(3):653. doi: 10.3390/v15030653. Viruses. 2023. PMID: 36992363 Free PMC article. Review.
-
In Silico Identification and In Vitro Validation of Repurposed Compounds Targeting the RSV Polymerase.Microorganisms. 2023 Jun 18;11(6):1608. doi: 10.3390/microorganisms11061608. Microorganisms. 2023. PMID: 37375110 Free PMC article.
References
-
- Lamb RA. 2013. Mononegavirales In Knipe DM, Howley PM (ed), Fields virology, 6th ed, Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chabi-Jesus C, Chandran K, Chiapponi C, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Griffiths A, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lavazza A, Lee B, Lelli D, Leroy EM, Lǐ J, Maes P, Marzano S-YL, Moreno A, Mühlberger E, Netesov SV, Nowotny N, Nylund A, et al. . 2019. Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. doi:10.1007/s00705-019-04247-4. - DOI - PMC - PubMed
-
- Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand F-X, Briese T, Bukreyev A, Calisher CH, Chandran K, Chéng J, Clawson AN, Collins PL, Dietzgen RG, Dolnik O, Domier LL, Dürrwald R, Dye JM, Easton AJ, Ebihara H, Farkas SL, Freitas-Astúa J, Formenty P, Fouchier RAM, Fù Y, Ghedin E, Goodin MM, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondo H, Kurath G, Lamb RA, Lenardon S, Leroy EM, Li C-X, Lin X-D, Liú L, Longdon B, Marton S, Maisner A, Mühlberger E, Netesov SV, Nowotny N, et al. . 2016. Taxonomy of the order Mononegavirales: update 2016. Arch Virol 161:2351–2360. doi:10.1007/s00705-016-2880-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

