SARS-CoV-2 and Three Related Coronaviruses Utilize Multiple ACE2 Orthologs and Are Potently Blocked by an Improved ACE2-Ig
- PMID: 32847856
- PMCID: PMC7592233
- DOI: 10.1128/JVI.01283-20
SARS-CoV-2 and Three Related Coronaviruses Utilize Multiple ACE2 Orthologs and Are Potently Blocked by an Improved ACE2-Ig
Abstract
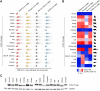
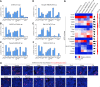
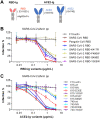
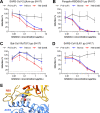
The ongoing coronavirus disease 2019 (COVID-19) pandemic has caused >20 million infections and >750,000 deaths. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, has been found closely related to the bat coronavirus strain RaTG13 (Bat-CoV RaTG13) and a recently identified pangolin coronavirus (Pangolin-CoV-2020). Here, we first investigated the ability of SARS-CoV-2 and three related coronaviruses to utilize animal orthologs of angiotensin-converting enzyme 2 (ACE2) for cell entry. We found that ACE2 orthologs of a wide range of domestic and wild mammals, including camels, cattle, horses, goats, sheep, cats, rabbits, and pangolins, were able to support cell entry of SARS-CoV-2, suggesting that these species might be able to harbor and spread this virus. In addition, the pangolin and bat coronaviruses, Pangolin-CoV-2020 and Bat-CoV RaTG13, were also found able to utilize human ACE2 and a number of animal-ACE2 orthologs for cell entry, indicating risks of spillover of these viruses into humans in the future. We then developed potently anticoronavirus ACE2-Ig proteins that are broadly effective against the four distinct coronaviruses. In particular, through truncating ACE2 at its residue 740 but not 615, introducing a D30E mutation, and adopting an antibody-like tetrameric-ACE2 configuration, we generated an ACE2-Ig variant that neutralizes SARS-CoV-2 at picomolar range. These data demonstrate that the improved ACE2-Ig variants developed in this study could potentially be developed to protect from SARS-CoV-2 and some other SARS-like viruses that might spillover into humans in the future.IMPORTANCE The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of the currently uncontrolled coronavirus disease 2019 (COVID-19) pandemic. It is important to study the host range of SARS-CoV-2, because some domestic species might harbor the virus and transmit it back to humans. In addition, insight into the ability of SARS-CoV-2 and SARS-like viruses to utilize animal orthologs of the SARS-CoV-2 receptor ACE2 might provide structural insight into improving ACE2-based viral entry inhibitors. In this study, we found that ACE2 orthologs of a wide range of domestic and wild animals can support cell entry of SARS-CoV-2 and three related coronaviruses, providing insights into identifying animal hosts of these viruses. We also developed recombinant ACE2-Ig proteins that are able to potently block these viral infections, providing a promising approach to developing antiviral proteins broadly effective against these distinct coronaviruses.
Keywords: ACE2; ACE2-Ig; SARS-CoV-2; entry inhibitor; host range.
Copyright © 2020 American Society for Microbiology.
Figures

Comment in
-
A binding-enhanced but enzymatic activity-eliminated human ACE2 efficiently neutralizes SARS-CoV-2 variants.Signal Transduct Target Ther. 2022 Jan 11;7(1):10. doi: 10.1038/s41392-021-00821-y. Signal Transduct Target Ther. 2022. PMID: 35013100 Free PMC article. No abstract available.
Similar articles
-
Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2.J Virol. 2020 Aug 31;94(18):e00940-20. doi: 10.1128/JVI.00940-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32661139 Free PMC article.
-
Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts.J Virol. 2020 Jul 16;94(15):e00831-20. doi: 10.1128/JVI.00831-20. Print 2020 Jul 16. J Virol. 2020. PMID: 32404529 Free PMC article.
-
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor.Cell. 2020 Apr 16;181(2):271-280.e8. doi: 10.1016/j.cell.2020.02.052. Epub 2020 Mar 5. Cell. 2020. PMID: 32142651 Free PMC article.
-
Coronaviruses and SARS-COV-2.Turk J Med Sci. 2020 Apr 21;50(SI-1):549-556. doi: 10.3906/sag-2004-127. Turk J Med Sci. 2020. PMID: 32293832 Free PMC article. Review.
-
[Source of the COVID-19 pandemic: ecology and genetics of coronaviruses (Betacoronavirus: Coronaviridae) SARS-CoV, SARS-CoV-2 (subgenus Sarbecovirus), and MERS-CoV (subgenus Merbecovirus).].Vopr Virusol. 2020;65(2):62-70. doi: 10.36233/0507-4088-2020-65-2-62-70. Vopr Virusol. 2020. PMID: 32515561 Review. Russian.
Cited by
-
Development and application of therapeutic antibodies against COVID-19.Int J Biol Sci. 2021 Apr 10;17(6):1486-1496. doi: 10.7150/ijbs.59149. eCollection 2021. Int J Biol Sci. 2021. PMID: 33907512 Free PMC article. Review.
-
Comparative analysis reveals the species-specific genetic determinants of ACE2 required for SARS-CoV-2 entry.PLoS Pathog. 2021 Mar 24;17(3):e1009392. doi: 10.1371/journal.ppat.1009392. eCollection 2021 Mar. PLoS Pathog. 2021. PMID: 33760889 Free PMC article.
-
Establishment of a human organoid-based evaluation system for assessing interspecies infection risk of animal-borne coronaviruses.Emerg Microbes Infect. 2024 Dec;13(1):2327368. doi: 10.1080/22221751.2024.2327368. Epub 2024 Mar 26. Emerg Microbes Infect. 2024. PMID: 38531008 Free PMC article.
-
Multifunctional angiotensin converting enzyme 2, the SARS-CoV-2 entry receptor, and critical appraisal of its role in acute lung injury.Biomed Pharmacother. 2021 Apr;136:111193. doi: 10.1016/j.biopha.2020.111193. Epub 2021 Jan 5. Biomed Pharmacother. 2021. PMID: 33461019 Free PMC article. Review.
-
A comprehensive guide to the pharmacologic regulation of angiotensin converting enzyme 2 (ACE2), the SARS-CoV-2 entry receptor.Pharmacol Ther. 2021 May;221:107750. doi: 10.1016/j.pharmthera.2020.107750. Epub 2020 Dec 1. Pharmacol Ther. 2021. PMID: 33275999 Free PMC article. Review.
References
-
- Zeng FY, Chan CW, Chan MN, Chen JD, Chow KY, Hon CC, Hui KH, Li J, Li VY, Wang CY, Wang PY, Guan Y, Zheng B, Poon LL, Chan KH, Yuen KY, Peiris JS, Leung FC. 2003. The complete genome sequence of severe acute respiratory syndrome coronavirus strain HKU-39849 (HK-39). Exp Biol Med (Maywood) 228:866–873. doi:10.1177/15353702-0322807-13. - DOI - PubMed
-
- Chim SS, Tsui SK, Chan KC, Au TC, Hung EC, Tong YK, Chiu RW, Ng EK, Chan PK, Chu CM, Sung JJ, Tam JS, Fung KP, Waye MM, Lee CY, Yuen KY, Lo YM, Group CMSR. 2003. Genomic characterization of the severe acute respiratory syndrome coronavirus of Amoy Gardens outbreak in Hong Kong. Lancet 362:1807–1808. doi:10.1016/S0140-6736(03)14901-X. - DOI - PMC - PubMed
-
- Zhong NS, Zheng BJ, Li YM, Poon LLM, Xie ZH, Chan KH, Li PH, Tan SY, Chang Q, Xie JP, Liu XQ, Xu J, Li DX, Yuen KY, Peiris, Guan Y. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362:1353–1358. doi:10.1016/S0140-6736(03)14630-2. - DOI - PMC - PubMed
-
- Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. doi:10.1038/s41586-018-0010-9. - DOI - PMC - PubMed
-
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi:10.1126/science.1087139. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

