Design, Synthesis, and Biological Evaluation of Novel 7 H-[1,2,4]Triazolo[3,4- b][1,3,4]thiadiazine Inhibitors as Antitumor Agents
- PMID: 32832771
- PMCID: PMC7439371
- DOI: 10.1021/acsomega.0c01829
Design, Synthesis, and Biological Evaluation of Novel 7 H-[1,2,4]Triazolo[3,4- b][1,3,4]thiadiazine Inhibitors as Antitumor Agents
Abstract
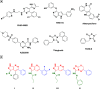
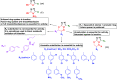
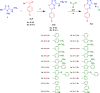
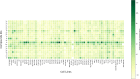
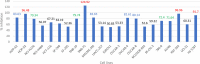
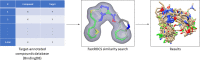
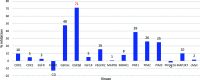
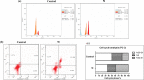
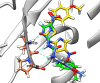
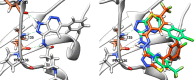
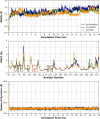
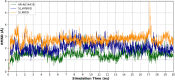
A series of novel anticancer hydrazinotriazolothiadiazine-based derivatives were designed based on the structure-activity relationship of the previously reported anticancer triazolothiadiazines. These derivatives were synthesized and biologically screened against full NCI-60 cancer cell lines revealing compound 5l with a potential antiproliferative effect. 5l was screened over 16 kinases to study its cytotoxic mechanism which showed to inhibit glycogen synthase kinase-3 β (GSK-3β) with IC50 equal to 0.883 μM and 14-fold selectivity over CDK2. Also, 5l increased active caspase-3 levels, induced cell cycle arrest at the G2-M phase, and increased the percentage of Annexin V-fluorescein isothiocyanate-positive apoptotic cells in PC-3 prostate cancer-treated cells. Molecular docking and dynamics were performed to predict the binding mode of 5l in the GSK-3β ATP binding site. 5l can be utilized as a starting scaffold for developing potential GSK-3β inhibitors.
Copyright © 2020 American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Vision on Synthetic and Medicinal Facets of 1,2,4-Triazolo[3,4-b][1,3,4]thiadiazine Scaffold.Top Curr Chem (Cham). 2022 Feb 5;380(2):10. doi: 10.1007/s41061-022-00365-x. Top Curr Chem (Cham). 2022. PMID: 35122161 Free PMC article. Review.
-
Design, synthesis and biological evaluation of N-alkyl or aryl substituted isoindigo derivatives as potential dual cyclin-dependent kinase 2 (CDK2)/glycogen synthase kinase 3β (GSK-3β) phosphorylation inhibitors.Eur J Med Chem. 2014 Oct 30;86:165-74. doi: 10.1016/j.ejmech.2014.08.049. Epub 2014 Aug 15. Eur J Med Chem. 2014. PMID: 25151579
-
Design, synthesis and structure-activity relationship of 3,6-diaryl-7H-[1,2,4]triazolo[3,4-b][1,3,4]thiadiazines as novel tubulin inhibitors.Sci Rep. 2017 Sep 20;7(1):11997. doi: 10.1038/s41598-017-10860-7. Sci Rep. 2017. PMID: 28931885 Free PMC article.
-
Novel Pyrazolo[3,4-d]pyrimidines as Potential Cytotoxic Agents: Design, Synthesis, Molecular Docking and CDK2 Inhibition.Anticancer Agents Med Chem. 2019;19(11):1368-1381. doi: 10.2174/1871520619666190417153350. Anticancer Agents Med Chem. 2019. PMID: 31038080
-
Drug development targeting the glycogen synthase kinase-3beta (GSK-3beta)-mediated signal transduction pathway: inhibitors of the Wnt/beta-catenin signaling pathway as novel anticancer drugs.J Pharmacol Sci. 2009 Feb;109(2):179-83. doi: 10.1254/jphs.08r28fm. Epub 2009 Jan 29. J Pharmacol Sci. 2009. PMID: 19179804 Review.
Cited by
-
Synthetic Methods and Pharmacological Potentials of Triazolothiadiazines: A Review.Molecules. 2024 Mar 16;29(6):1326. doi: 10.3390/molecules29061326. Molecules. 2024. PMID: 38542962 Free PMC article. Review.
-
Convenient synthesis and X-ray determination of 2-amino-6H-1,3,4-thiadiazin-3-ium bromides endowed with antiproliferative activity.RSC Adv. 2024 Jun 27;14(25):17866-17876. doi: 10.1039/d4ra02531h. eCollection 2024 May 28. RSC Adv. 2024. PMID: 38939040 Free PMC article.
-
Vision on Synthetic and Medicinal Facets of 1,2,4-Triazolo[3,4-b][1,3,4]thiadiazine Scaffold.Top Curr Chem (Cham). 2022 Feb 5;380(2):10. doi: 10.1007/s41061-022-00365-x. Top Curr Chem (Cham). 2022. PMID: 35122161 Free PMC article. Review.
-
Synthesis and Anti-Breast Cancer Potency of Mono- and Bis-(pyrazolyl[1,2,4]triazolo[3,4-b][1,3,4]thiadiazine) Derivatives as EGFR/CDK-2 Target Inhibitors.ACS Omega. 2023 Sep 11;8(38):35359-35369. doi: 10.1021/acsomega.3c05309. eCollection 2023 Sep 26. ACS Omega. 2023. PMID: 37779952 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

