SirT7 auto-ADP-ribosylation regulates glucose starvation response through mH2A1
- PMID: 32832656
- PMCID: PMC7439345
- DOI: 10.1126/sciadv.aaz2590
SirT7 auto-ADP-ribosylation regulates glucose starvation response through mH2A1
Abstract
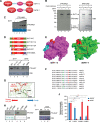
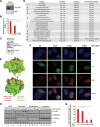
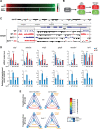
Sirtuins are key players of metabolic stress response. Originally described as deacetylases, some sirtuins also exhibit poorly understood mono-adenosine 5'-diphosphate (ADP)-ribosyltransferase (mADPRT) activity. We report that the deacetylase SirT7 is a dual sirtuin, as it also features auto-mADPRT activity. SirT7 mADPRT occurs at a previously undefined active site, and its abrogation alters SirT7 chromatin distribution. We identify an epigenetic pathway by which ADP-ribosyl-SirT7 is recognized by the ADP-ribose reader mH2A1.1 under glucose starvation, inducing SirT7 relocalization to intergenic regions. SirT7 promotes mH2A1 enrichment in a subset of nearby genes, many of them involved in second messenger signaling, resulting in their specific up- or down-regulation. The expression profile of these genes under calorie restriction is consistently abrogated in SirT7-deficient mice, resulting in impaired activation of autophagy. Our work provides a novel perspective on sirtuin duality and suggests a role for SirT7/mH2A1.1 axis in glucose homeostasis and aging.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures



Similar articles
-
Biochemical characterization of mono ADP ribosyl transferase activity of human sirtuin SIRT7 and its regulation.Arch Biochem Biophys. 2020 Feb 15;680:108226. doi: 10.1016/j.abb.2019.108226. Epub 2019 Dec 13. Arch Biochem Biophys. 2020. PMID: 31843644
-
SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability.Nat Commun. 2016 Jul 20;7:12235. doi: 10.1038/ncomms12235. Nat Commun. 2016. PMID: 27436229 Free PMC article.
-
Sirtuin 7 in cell proliferation, stress and disease: Rise of the Seventh Sirtuin!Cell Signal. 2015 Mar;27(3):673-82. doi: 10.1016/j.cellsig.2014.11.026. Epub 2014 Nov 27. Cell Signal. 2015. PMID: 25435428 Review.
-
Repairing split ends: SIRT6, mono-ADP ribosylation and DNA repair.Aging (Albany NY). 2011 Sep;3(9):829-35. doi: 10.18632/aging.100389. Aging (Albany NY). 2011. PMID: 21946623 Free PMC article. Review.
-
SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context.ACS Chem Biol. 2016 Mar 18;11(3):742-7. doi: 10.1021/acschembio.5b01084. Epub 2016 Mar 3. ACS Chem Biol. 2016. PMID: 26907567 Free PMC article.
Cited by
-
Multifunctional histone variants in genome function.Nat Rev Genet. 2024 Aug 13. doi: 10.1038/s41576-024-00759-1. Online ahead of print. Nat Rev Genet. 2024. PMID: 39138293 Review.
-
Sirtuin-dependent metabolic and epigenetic regulation of macrophages during tuberculosis.Front Immunol. 2023 Mar 13;14:1121495. doi: 10.3389/fimmu.2023.1121495. eCollection 2023. Front Immunol. 2023. PMID: 36993975 Free PMC article. Review.
-
SIRT7: a novel molecular target for personalized cancer treatment?Oncogene. 2024 Mar;43(14):993-1006. doi: 10.1038/s41388-024-02976-8. Epub 2024 Feb 21. Oncogene. 2024. PMID: 38383727 Free PMC article. Review.
-
The Emerging Role of SIRT7 in Glucose and Lipid Metabolism.Cells. 2023 Dec 25;13(1):48. doi: 10.3390/cells13010048. Cells. 2023. PMID: 38201252 Free PMC article. Review.
-
The Role of MacroH2A Histone Variants in Cancer.Cancers (Basel). 2021 Jun 15;13(12):3003. doi: 10.3390/cancers13123003. Cancers (Basel). 2021. PMID: 34203934 Free PMC article. Review.
References
-
- Bosch-Presegué L., Vaquero A., Sirtuins in stress response: Guardians of the genome. Oncogene 33, 3764–3775 (2014). - PubMed
-
- Caban C. E., Ginsburg A., Glutamine synthetase adenylyltransferase from Escherichia coli: Purification and physical and chemical properties. Biochemistry 15, 1569–1580 (1976). - PubMed
-
- Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S., Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 (1998). - PubMed

