LIN28B/ let-7 control the ability of neonatal murine auditory supporting cells to generate hair cells through mTOR signaling
- PMID: 32826333
- PMCID: PMC7486708
- DOI: 10.1073/pnas.2000417117
LIN28B/ let-7 control the ability of neonatal murine auditory supporting cells to generate hair cells through mTOR signaling
Abstract
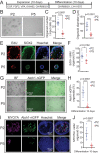
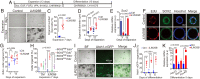
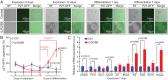
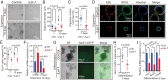
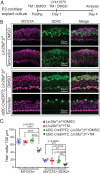
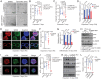
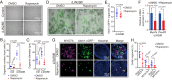
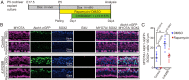
Mechano-sensory hair cells within the inner ear cochlea are essential for the detection of sound. In mammals, cochlear hair cells are only produced during development and their loss, due to disease or trauma, is a leading cause of deafness. In the immature cochlea, prior to the onset of hearing, hair cell loss stimulates neighboring supporting cells to act as hair cell progenitors and produce new hair cells. However, for reasons unknown, such regenerative capacity (plasticity) is lost once supporting cells undergo maturation. Here, we demonstrate that the RNA binding protein LIN28B plays an important role in the production of hair cells by supporting cells and provide evidence that the developmental drop in supporting cell plasticity in the mammalian cochlea is, at least in part, a product of declining LIN28B-mammalian target of rapamycin (mTOR) activity. Employing murine cochlear organoid and explant cultures to model mitotic and nonmitotic mechanisms of hair cell generation, we show that loss of LIN28B function, due to its conditional deletion, or due to overexpression of the antagonistic miRNA let-7g, suppressed Akt-mTOR complex 1 (mTORC1) activity and renders young, immature supporting cells incapable of generating hair cells. Conversely, we found that LIN28B overexpression increased Akt-mTORC1 activity and allowed supporting cells that were undergoing maturation to de-differentiate into progenitor-like cells and to produce hair cells via mitotic and nonmitotic mechanisms. Finally, using the mTORC1 inhibitor rapamycin, we demonstrate that LIN28B promotes supporting cell plasticity in an mTORC1-dependent manner.
Keywords: LIN28; hair cell regeneration; inner ear cochlea; let-7 miRNA; mTOR pathway.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
The RNA-binding protein LIN28B regulates developmental timing in the mammalian cochlea.Proc Natl Acad Sci U S A. 2015 Jul 21;112(29):E3864-73. doi: 10.1073/pnas.1501077112. Epub 2015 Jul 2. Proc Natl Acad Sci U S A. 2015. PMID: 26139524 Free PMC article.
-
Follistatin promotes LIN28B-mediated supporting cell reprogramming and hair cell regeneration in the murine cochlea.Sci Adv. 2022 Feb 11;8(6):eabj7651. doi: 10.1126/sciadv.abj7651. Epub 2022 Feb 11. Sci Adv. 2022. PMID: 35148175 Free PMC article.
-
TRIM71 reactivation enhances the mitotic and hair cell-forming potential of cochlear supporting cells.EMBO Rep. 2023 Sep 6;24(9):e56562. doi: 10.15252/embr.202256562. Epub 2023 Jul 26. EMBO Rep. 2023. PMID: 37492931 Free PMC article.
-
Hair cell regeneration in the avian auditory epithelium.Int J Dev Biol. 2007;51(6-7):633-47. doi: 10.1387/ijdb.072408js. Int J Dev Biol. 2007. PMID: 17891722 Review.
-
Role of Wnt and Notch signaling in regulating hair cell regeneration in the cochlea.Front Med. 2016 Sep;10(3):237-49. doi: 10.1007/s11684-016-0464-9. Epub 2016 Sep 7. Front Med. 2016. PMID: 27527363 Review.
Cited by
-
Hair Cell Regeneration: From Animals to Humans.Clin Exp Otorhinolaryngol. 2024 Feb;17(1):1-14. doi: 10.21053/ceo.2023.01382. Epub 2024 Jan 19. Clin Exp Otorhinolaryngol. 2024. PMID: 38271988 Free PMC article. Review.
-
Stem Cell-Based Hair Cell Regeneration and Therapy in the Inner Ear.Neurosci Bull. 2024 Jan;40(1):113-126. doi: 10.1007/s12264-023-01130-w. Epub 2023 Oct 3. Neurosci Bull. 2024. PMID: 37787875 Free PMC article. Review.
-
A Low Dose of Rapamycin Promotes Hair Cell Differentiation by Enriching SOX2+ Progenitors in the Neonatal Mouse Inner Ear Organoids.J Assoc Res Otolaryngol. 2024 Apr;25(2):149-165. doi: 10.1007/s10162-024-00938-1. Epub 2024 Mar 12. J Assoc Res Otolaryngol. 2024. PMID: 38472516
-
Future Pharmacotherapy for Sensorineural Hearing Loss by Protection and Regeneration of Auditory Hair Cells.Pharmaceutics. 2023 Feb 26;15(3):777. doi: 10.3390/pharmaceutics15030777. Pharmaceutics. 2023. PMID: 36986638 Free PMC article. Review.
-
The Suppression of Ubiquitin C-Terminal Hydrolase L1 Promotes the Transdifferentiation of Auditory Supporting Cells into Hair Cells by Regulating the mTOR Pathway.Cells. 2024 Apr 24;13(9):737. doi: 10.3390/cells13090737. Cells. 2024. PMID: 38727276 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

