Continuous Reassortment of Clade 2.3.4.4 H5N6 Highly Pathogenetic Avian Influenza Viruses Demonstrating High Risk to Public Health
- PMID: 32824873
- PMCID: PMC7460007
- DOI: 10.3390/pathogens9080670
Continuous Reassortment of Clade 2.3.4.4 H5N6 Highly Pathogenetic Avian Influenza Viruses Demonstrating High Risk to Public Health
Abstract
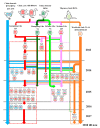
Since it firstly emerged in China in 2013, clade 2.3.4.4 H5N6 highly pathogenic avian influenza viruses (HPAIVs) has rapidly replaced predominant H5N1 to become the dominant H5 subtype in China, especially in ducks. Not only endemic in China, it also crossed the geographical barrier and emerged in South Korea, Japan, and Europe. Here, we analyzed the genetic properties of the clade 2.3.4.4 H5N6 HPAIVs with full genome sequences available online together with our own isolates. Phylogenetic analysis showed that clade 2.3.4.4 H5N6 HPAIVs continuously reassorted with local H5, H6, and H7N9/H9N2. Species analysis reveals that aquatic poultry and migratory birds became the dominant hosts of H5N6. Adaption to aquatic poultry might help clade 2.3.4.4 H5N6 better adapt to migratory birds, thus enabling it to become endemic in China. Besides, migratory birds might help clade 2.3.4.4 H5N6 transmit all over the world. Clade 2.3.4.4 H5N6 HPAIVs also showed a preference for α2,6-SA receptors when compared to other avian origin influenza viruses. Experiments in vitro and in vivo revealed that clade 2.3.4.4 H5N6 HPAIVs exhibited high replication efficiency in both avian and mammal cells, and it also showed high pathogenicity in both mice and chickens, demonstrating high risk to public health. Considering all the factors together, adaption to aquatic poultry and migratory birds helps clade 2.3.4.4 H5N6 overcome the geographical isolation, and it has potential to be the next influenza pandemic in the world, making it worthy of our attention.
Keywords: clade 2.3.4.4 H5N6; influenza virus; pandemic; pathogenicity; reassort; transmit.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Genetic analysis and biological characteristics of different internal gene origin H5N6 reassortment avian influenza virus in China in 2016.Vet Microbiol. 2018 Jun;219:200-211. doi: 10.1016/j.vetmic.2018.04.023. Epub 2018 Apr 18. Vet Microbiol. 2018. PMID: 29778197
-
Genesis and Dissemination of Highly Pathogenic H5N6 Avian Influenza Viruses.J Virol. 2017 Feb 14;91(5):e02199-16. doi: 10.1128/JVI.02199-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28003485 Free PMC article.
-
The Biological Characteristics of Novel H5N6 Highly Pathogenic Avian Influenza Virus and Its Pathogenesis in Ducks.Front Microbiol. 2021 Jan 26;12:628545. doi: 10.3389/fmicb.2021.628545. eCollection 2021. Front Microbiol. 2021. PMID: 33584629 Free PMC article.
-
Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4.J Vet Sci. 2017 Aug 31;18(S1):269-280. doi: 10.4142/jvs.2017.18.S1.269. J Vet Sci. 2017. PMID: 28859267 Free PMC article. Review.
-
Intracontinental and intercontinental dissemination of Asian H5 highly pathogenic avian influenza virus (clade 2.3.4.4) in the winter of 2014-2015.Rev Med Virol. 2015 Nov;25(6):388-405. doi: 10.1002/rmv.1857. Epub 2015 Oct 13. Rev Med Virol. 2015. PMID: 26458727 Review.
Cited by
-
Advances in Development and Application of Influenza Vaccines.Front Immunol. 2021 Jul 13;12:711997. doi: 10.3389/fimmu.2021.711997. eCollection 2021. Front Immunol. 2021. PMID: 34326849 Free PMC article. Review.
-
Phylogenetic and Phylogeographic Analysis of the Highly Pathogenic H5N6 Avian Influenza Virus in China.Viruses. 2022 Aug 11;14(8):1752. doi: 10.3390/v14081752. Viruses. 2022. PMID: 36016374 Free PMC article.
-
Successful combinatorial therapy of sirolimus and neuraminidase inhibitors in a patient with highly pathogenic avian influenza A (H5N6) virus: a case report.Ann Transl Med. 2022 Mar;10(5):265. doi: 10.21037/atm-22-704. Ann Transl Med. 2022. PMID: 35402590 Free PMC article.
-
The Alarming Situation of Highly Pathogenic Avian Influenza Viruses in 2019-2023.Glob Med Genet. 2024 Jun 28;11(3):200-213. doi: 10.1055/s-0044-1788039. eCollection 2024 Sep. Glob Med Genet. 2024. PMID: 38947761 Free PMC article. Review.
-
Progress in the Development of Universal Influenza Vaccines.Viruses. 2020 Sep 17;12(9):1033. doi: 10.3390/v12091033. Viruses. 2020. PMID: 32957468 Free PMC article. Review.
References
-
- Nguyen D.T., Jang Y., Nguyen T.D., Jones J., Shepard S.S., Yang H., Gerloff N., Fabrizio T., Nguyen L.V., Inui K., et al. Shifting Clade Distribution, Reassortment, and Emergence of New Subtypes of Highly Pathogenic Avian Influenza A(H5) Viruses Collected from Vietnamese Poultry from 2012 to 2015. J. Virol. 2017;91 doi: 10.1128/JVI.01708-16. - DOI - PMC - PubMed
-
- Kwon J.H., Lee D.H., Swayne D.E., Noh J.Y., Yuk S.S., Erdene-Ochir T.O., Hong W.T., Jeong J.H., Jeong S., Gwon G.B., et al. Reassortant Clade 2.3.4.4 Avian Influenza A(H5N6) Virus in a Wild Mandarin Duck, South Korea, 2016. Emerg. Infect. Dis. 2017;23:822–826. doi: 10.3201/eid2305.161905. - DOI - PMC - PubMed
Grants and funding
- 31830097/National Natural Science Foundation of China
- 31672586/National Natural Science Foundation of China
- 2019B020218004/Key Research and Development Program of Guangdong Province
- CARS-41-G16/Earmarked Found for China Agriculture Research System
- 2018, Wenbao Qi/Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme
LinkOut - more resources
Full Text Sources

