Proposal of De Novo Antigen Test for COVID-19: Ultrasensitive Detection of Spike Proteins of SARS-CoV-2
- PMID: 32823866
- PMCID: PMC7459804
- DOI: 10.3390/diagnostics10080594
Proposal of De Novo Antigen Test for COVID-19: Ultrasensitive Detection of Spike Proteins of SARS-CoV-2
Abstract
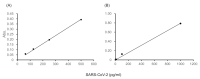
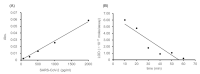
Polymerase chain reaction (PCR)-based antigen tests are technically difficult, time-consuming, and expensive, and may produce false negative results requiring follow-up confirmation with computed tomography. The global coronavirus disease 2019 (COVID-19) pandemic has increased the demand for accurate, easy-to-use, rapid, and cost-effective antigen tests for clinical application. We propose a de novo antigen test for diagnosing COVID-19 using the combination of sandwich enzyme-linked immunosorbent assay and thio-nicotinamide adenine dinucleotide (thio-NAD) cycling. Our test takes advantage of the spike proteins specific to the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus. The limit of detection of our test was 2.3 × 10-18 moles/assay. If the virus has ~25 spike proteins on its surface, our method should detect on the order of 10-20 moles of virus/assay, corresponding to ~104 copies of the virus RNA/assay. The detection sensitivity approaches that of PCR-based assays because the average virus RNA load used for PCR-based assays is ~105 copies per oro- or naso-pharyngeal swab specimen. To our knowledge, this is the first ultrasensitive antigen test for SARS-CoV-2 spike proteins that can be performed with an easy-to-use microplate reader. Sufficient sensitivity can be achieved within 10 min of thio-NAD cycling. Our antigen test allows for rapid, cost-effective, specific, ultrasensitive, and simultaneous multiple measurements of SARS-CoV-2, and has broad application for the diagnosis for COVID-19.
Keywords: COVID-19; SARS-CoV-2; spike protein; thio-NAD cycling; ultrasensitive ELISA.
Conflict of interest statement
R.T., N.A., K.N., and S.W. are employees of TAUNS Laboratories, Inc. E.I. received research expenses and a patent royalty from TAUNS Laboratories, Inc. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article. Review.
-
Ultrasensitive Detection of SARS-CoV-2 Spike Proteins Using the Thio-NAD Cycling Reaction: A Preliminary Study before Clinical Trials.Microorganisms. 2021 Oct 25;9(11):2214. doi: 10.3390/microorganisms9112214. Microorganisms. 2021. PMID: 34835340 Free PMC article.
-
Improved Detection Sensitivity of an Antigen Test for SARS-CoV-2 Nucleocapsid Proteins with Thio-NAD Cycling.Biol Pharm Bull. 2021 Sep 1;44(9):1332-1336. doi: 10.1248/bpb.b21-00387. Epub 2021 Jun 19. Biol Pharm Bull. 2021. PMID: 34148926
-
A Generic, Scalable, and Rapid Time-Resolved Förster Resonance Energy Transfer-Based Assay for Antigen Detection-SARS-CoV-2 as a Proof of Concept.mBio. 2021 May 18;12(3):e00902-21. doi: 10.1128/mBio.00902-21. mBio. 2021. PMID: 34006662 Free PMC article.
-
Ultrasensitive ELISA Developed for Diagnosis.Diagnostics (Basel). 2019 Jul 18;9(3):78. doi: 10.3390/diagnostics9030078. Diagnostics (Basel). 2019. PMID: 31323782 Free PMC article. Review.
Cited by
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article. Review.
-
Current Technologies for Detection of COVID-19: Biosensors, Artificial Intelligence and Internet of Medical Things (IoMT): Review.Sensors (Basel). 2022 Dec 30;23(1):426. doi: 10.3390/s23010426. Sensors (Basel). 2022. PMID: 36617023 Free PMC article. Review.
-
Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2021 Mar 24;3(3):CD013705. doi: 10.1002/14651858.CD013705.pub2. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 Jul 22;7:CD013705. doi: 10.1002/14651858.CD013705.pub3. PMID: 33760236 Free PMC article. Updated.
-
Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics.Lab Invest. 2022 Jan;102(1):4-13. doi: 10.1038/s41374-021-00663-w. Epub 2021 Sep 8. Lab Invest. 2022. PMID: 34497366 Free PMC article. Review.
-
Zeptomole Detection of an Enzyme by a Simple Colorimetric Method.Anal Sci. 2021 Oct 10;37(10):1469-1472. doi: 10.2116/analsci.21N009. Epub 2021 Mar 19. Anal Sci. 2021. PMID: 33746140
References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- Naming the Coronavirus Disease (COVID-19) and the Virus that Causes It. [(accessed on 12 July 2020)]; Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technica....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

