Coxiella burnetii-Infected NK Cells Release Infectious Bacteria by Degranulation
- PMID: 32817330
- PMCID: PMC7573450
- DOI: 10.1128/IAI.00172-20
Coxiella burnetii-Infected NK Cells Release Infectious Bacteria by Degranulation
Abstract
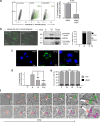
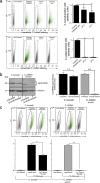
Natural killer (NK) cells are critically involved in the early immune response against various intracellular pathogens, including Coxiella burnetii and Chlamydia psittaciChlamydia-infected NK cells functionally mature, induce cellular immunity, and protect themselves by killing the bacteria in secreted granules. Here, we report that infected NK cells do not allow intracellular multiday growth of Coxiella, as is usually observed in other host cell types. C. burnetii-infected NK cells display maturation and gamma interferon (IFN-γ) secretion, as well as the release of Coxiella-containing lytic granules. Thus, NK cells possess a potent program to restrain and expel different types of invading bacteria via degranulation. Strikingly, though, in contrast to Chlamydia, expulsed Coxiella organisms largely retain their infectivity and, hence, escape the cell-autonomous self-defense mechanism in NK cells.
Keywords: Chlamydia psittaci; Coxiella burnetii; NK cells; cell-autonomous immunity.
Copyright © 2020 American Society for Microbiology.
Figures





Similar articles
-
TGF-β/IFN-γ Antagonism in Subversion and Self-Defense of Phase II Coxiella burnetii-Infected Dendritic Cells.Infect Immun. 2023 Feb 16;91(2):e0032322. doi: 10.1128/iai.00323-22. Epub 2023 Jan 23. Infect Immun. 2023. PMID: 36688662 Free PMC article. Clinical Trial.
-
NK Cell-Mediated Processing Of Chlamydia psittaci Drives Potent Anti-Bacterial Th1 Immunity.Sci Rep. 2019 Mar 18;9(1):4799. doi: 10.1038/s41598-019-41264-4. Sci Rep. 2019. PMID: 30886314 Free PMC article.
-
Early cytokine and antibody responses against Coxiella burnetii in aerosol infection of BALB/c mice.Diagn Microbiol Infect Dis. 2015 Apr;81(4):234-9. doi: 10.1016/j.diagmicrobio.2014.12.008. Epub 2014 Dec 30. Diagn Microbiol Infect Dis. 2015. PMID: 25618420 Free PMC article.
-
Adaptive immunity to the obligate intracellular pathogen Coxiella burnetii.Immunol Res. 2009;43(1-3):138-48. doi: 10.1007/s12026-008-8059-4. Immunol Res. 2009. PMID: 18813881 Free PMC article. Review.
-
Coxiella burnetii Lipopolysaccharide: What Do We Know?Int J Mol Sci. 2017 Nov 23;18(12):2509. doi: 10.3390/ijms18122509. Int J Mol Sci. 2017. PMID: 29168790 Free PMC article. Review.
Cited by
-
Macrophages inhibit Coxiella burnetii by the ACOD1-itaconate pathway for containment of Q fever.EMBO Mol Med. 2023 Feb 8;15(2):e15931. doi: 10.15252/emmm.202215931. Epub 2022 Dec 7. EMBO Mol Med. 2023. PMID: 36479617 Free PMC article.
-
Exploring epigenetic reprogramming during central nervous system infection.Immunol Rev. 2022 Oct;311(1):112-129. doi: 10.1111/imr.13079. Epub 2022 Apr 28. Immunol Rev. 2022. PMID: 35481573 Free PMC article. Review.
-
TGF-β/IFN-γ Antagonism in Subversion and Self-Defense of Phase II Coxiella burnetii-Infected Dendritic Cells.Infect Immun. 2023 Feb 16;91(2):e0032322. doi: 10.1128/iai.00323-22. Epub 2023 Jan 23. Infect Immun. 2023. PMID: 36688662 Free PMC article. Clinical Trial.
-
QuilA® adjuvanted Coxevac® sustains Th1-CD8+-type immunity and increases protection in Coxiella burnetii-challenged goats.NPJ Vaccines. 2023 Feb 14;8(1):17. doi: 10.1038/s41541-023-00607-z. NPJ Vaccines. 2023. PMID: 36788233 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

