Cryptic genetic variation enhances primate L1 retrotransposon survival by enlarging the functional coiled coil sequence space of ORF1p
- PMID: 32797042
- PMCID: PMC7449397
- DOI: 10.1371/journal.pgen.1008991
Cryptic genetic variation enhances primate L1 retrotransposon survival by enlarging the functional coiled coil sequence space of ORF1p
Abstract
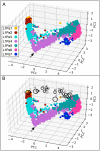
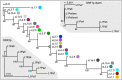
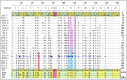
Accounting for continual evolution of deleterious L1 retrotransposon families, which can contain hundreds to thousands of members remains a major issue in mammalian biology. L1 activity generated upwards of 40% of some mammalian genomes, including humans where they remain active, causing genetic defects and rearrangements. L1 encodes a coiled coil-containing protein that is essential for retrotransposition, and the emergence of novel primate L1 families has been correlated with episodes of extensive amino acid substitutions in the coiled coil. These results were interpreted as an adaptive response to maintain L1 activity, however its mechanism remained unknown. Although an adventitious mutation can inactivate coiled coil function, its effect could be buffered by epistatic interactions within the coiled coil, made more likely if the family contains a diverse set of coiled coil sequences-collectively referred to as the coiled coil sequence space. Amino acid substitutions that do not affect coiled coil function (i.e., its phenotype) could be "hidden" from (not subject to) purifying selection. The accumulation of such substitutions, often referred to as cryptic genetic variation, has been documented in various proteins. Here we report that this phenomenon was in effect during the latest episode of primate coiled coil evolution, which occurred 30-10 MYA during the emergence of primate L1Pa7-L1Pa3 families. First, we experimentally demonstrated that while coiled coil function (measured by retrotransposition) can be eliminated by single epistatic mutations, it nonetheless can also withstand extensive amino acid substitutions. Second, principal component and cluster analysis showed that the coiled coil sequence space of each of the L1Pa7-3 families was notably increased by the presence of distinct, coexisting coiled coil sequences. Thus, sampling related networks of functional sequences rather than traversing discrete adaptive states characterized the persistence L1 activity during this evolutionary event.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Co-expression of distinct L1 retrotransposon coiled coils can lead to their entanglement.Mob DNA. 2023 Oct 20;14(1):16. doi: 10.1186/s13100-023-00303-8. Mob DNA. 2023. PMID: 37864180 Free PMC article.
-
L1 retrotransposition requires rapid ORF1p oligomerization, a novel coiled coil-dependent property conserved despite extensive remodeling.Nucleic Acids Res. 2016 Jan 8;44(1):281-93. doi: 10.1093/nar/gkv1342. Epub 2015 Dec 15. Nucleic Acids Res. 2016. PMID: 26673717 Free PMC article.
-
Condensation of LINE-1 is critical for retrotransposition.Elife. 2023 Apr 28;12:e82991. doi: 10.7554/eLife.82991. Elife. 2023. PMID: 37114770 Free PMC article.
-
Protein-nucleic acid interactions of LINE-1 ORF1p.Semin Cell Dev Biol. 2019 Feb;86:140-149. doi: 10.1016/j.semcdb.2018.03.019. Epub 2018 Mar 31. Semin Cell Dev Biol. 2019. PMID: 29596909 Free PMC article. Review.
-
LINE drive. retrotransposition and genome instability.Cell. 2002 Aug 9;110(3):277-80. doi: 10.1016/s0092-8674(02)00868-1. Cell. 2002. PMID: 12176313 Review.
Cited by
-
Reconstruction of full-length LINE-1 progenitors from ancestral genomes.Genetics. 2022 Jul 4;221(3):iyac074. doi: 10.1093/genetics/iyac074. Genetics. 2022. PMID: 35552404 Free PMC article.
-
Somatic retrotransposition in the developing rhesus macaque brain.Genome Res. 2022 Jul;32(7):1298-1314. doi: 10.1101/gr.276451.121. Epub 2022 Jun 21. Genome Res. 2022. PMID: 35728967 Free PMC article.
-
Co-expression of distinct L1 retrotransposon coiled coils can lead to their entanglement.Mob DNA. 2023 Oct 20;14(1):16. doi: 10.1186/s13100-023-00303-8. Mob DNA. 2023. PMID: 37864180 Free PMC article.
-
The L1-ORF1p coiled coil enables formation of a tightly compacted nucleic acid-bound complex that is associated with retrotransposition.Nucleic Acids Res. 2022 Aug 26;50(15):8690-8699. doi: 10.1093/nar/gkac628. Nucleic Acids Res. 2022. PMID: 35871298 Free PMC article.
-
Investigating the phylogenetic history of toxin tolerance in mushroom-feeding Drosophila.Ecol Evol. 2023 Dec 13;13(12):e10736. doi: 10.1002/ece3.10736. eCollection 2023 Dec. Ecol Evol. 2023. PMID: 38099137 Free PMC article.
References
-
- IHGS-Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

