Genomic characterization of malignant progression in neoplastic pancreatic cysts
- PMID: 32796935
- PMCID: PMC7428044
- DOI: 10.1038/s41467-020-17917-8
Genomic characterization of malignant progression in neoplastic pancreatic cysts
Abstract
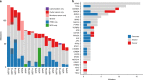
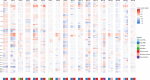
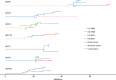
Intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs) are non-invasive neoplasms that are often observed in association with invasive pancreatic cancers, but their origins and evolutionary relationships are poorly understood. In this study, we analyze 148 samples from IPMNs, MCNs, and small associated invasive carcinomas from 18 patients using whole exome or targeted sequencing. Using evolutionary analyses, we establish that both IPMNs and MCNs are direct precursors to pancreatic cancer. Mutations in SMAD4 and TGFBR2 are frequently restricted to invasive carcinoma, while RNF43 alterations are largely in non-invasive lesions. Genomic analyses suggest an average window of over three years between the development of high-grade dysplasia and pancreatic cancer. Taken together, these data establish non-invasive IPMNs and MCNs as origins of invasive pancreatic cancer, identifying potential drivers of invasion, highlighting the complex clonal dynamics prior to malignant transformation, and providing opportunities for early detection and intervention.
Conflict of interest statement
L.D.W. receives research funding from Applied Materials. V.E.V. is a founder of Personal Genome Diagnostics, a member of its Scientific Advisory Board and Board of Directors, and owns Personal Genome Diagnostics stock, which are subject to certain restrictions under university policy. V.E.V. is an advisor to Takeda Pharmaceuticals. Within the last five years, V.E.V. has been an advisor to Daiichi Sankyo, Janssen Diagnostics, and Ignyta. J.R.W. is founder and owner of Resphera Biosciences LLC, and is a consultant to Personal Genome Diagnostics Inc. The terms of these arrangements are managed by Johns Hopkins University in accordance with its conflict of interest policies. The other authors declare no conflict of interest.
Figures

Similar articles
-
Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways.Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21188-93. doi: 10.1073/pnas.1118046108. Epub 2011 Dec 8. Proc Natl Acad Sci U S A. 2011. PMID: 22158988 Free PMC article.
-
Neoplastic Progression in Macroscopic Precursor Lesions of the Pancreas.Arch Pathol Lab Med. 2024 Sep 1;148(9):980-988. doi: 10.5858/arpa.2023-0358-RA. Arch Pathol Lab Med. 2024. PMID: 38386006 Review.
-
Intraductal Papillary Mucinous Neoplasms Arise From Multiple Independent Clones, Each With Distinct Mutations.Gastroenterology. 2019 Oct;157(4):1123-1137.e22. doi: 10.1053/j.gastro.2019.06.001. Epub 2019 Jun 5. Gastroenterology. 2019. PMID: 31175866 Free PMC article.
-
Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas.J Pathol. 2014 Jul;233(3):217-27. doi: 10.1002/path.4344. J Pathol. 2014. PMID: 24604757 Free PMC article.
-
Molecular pathways in pancreatic carcinogenesis.J Surg Oncol. 2013 Jan;107(1):8-14. doi: 10.1002/jso.23213. Epub 2012 Jul 17. J Surg Oncol. 2013. PMID: 22806689 Free PMC article. Review.
Cited by
-
Loss of Rnf43 Accelerates Kras-Mediated Neoplasia and Remodels the Tumor Immune Microenvironment in Pancreatic Adenocarcinoma.Gastroenterology. 2022 Apr;162(4):1303-1318.e18. doi: 10.1053/j.gastro.2021.12.273. Epub 2021 Dec 30. Gastroenterology. 2022. PMID: 34973294 Free PMC article.
-
A Review of the Diagnosis and Management of Premalignant Pancreatic Cystic Lesions.J Clin Med. 2021 Mar 19;10(6):1284. doi: 10.3390/jcm10061284. J Clin Med. 2021. PMID: 33808853 Free PMC article. Review.
-
Neoplastic cell enrichment of tumor tissues using coring and laser microdissection for proteomic and genomic analyses of pancreatic ductal adenocarcinoma.Clin Proteomics. 2022 Oct 20;19(1):36. doi: 10.1186/s12014-022-09373-x. Clin Proteomics. 2022. PMID: 36266629 Free PMC article.
-
Surgical treatment for pancreatic cystic lesions-implications from the multi-center and prospective German StuDoQ|Pancreas registry.Langenbecks Arch Surg. 2023 Jan 14;408(1):28. doi: 10.1007/s00423-022-02740-0. Langenbecks Arch Surg. 2023. PMID: 36640188 Free PMC article.
-
The Role of Genetic, Metabolic, Inflammatory, and Immunologic Mediators in the Progression of Intraductal Papillary Mucinous Neoplasms to Pancreatic Adenocarcinoma.Cancers (Basel). 2023 Mar 11;15(6):1722. doi: 10.3390/cancers15061722. Cancers (Basel). 2023. PMID: 36980608 Free PMC article. Review.
References
-
- Rahib L, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. - PubMed
-
- Lermite E, et al. Complications after pancreatic resection: diagnosis, prevention and management. Clin. Res. Hepatol. Gastroenterol. 2013;37:230–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

