Biphasic Liquid Microjunction Extraction for Profiling Neuronal RNA Modifications by Liquid Chromatography-Tandem Mass Spectrometry
- PMID: 32786436
- PMCID: PMC7496823
- DOI: 10.1021/acs.analchem.0c02830
Biphasic Liquid Microjunction Extraction for Profiling Neuronal RNA Modifications by Liquid Chromatography-Tandem Mass Spectrometry
Abstract
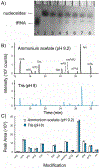
RNA modifications are emerging as critical players in the spatiotemporal regulation of gene expression. Although liquid chromatography-tandem mass spectrometry (LC-MS/MS) enables the simultaneous quantification of numerous enzymatically modified RNAs in a biological sample, conventional RNA extraction and enzymatic digestion protocols that are employed prior to analysis have precluded the application of this technique for small-volume samples. In this study, a biphasic liquid microjunction (LMJ) extraction system using coaxial capillaries that direct and aspirate extraction solvents onto a ∼350 μm diameter sample spot was developed and applied for the extraction of RNA from individual cell clusters in the central nervous system of the marine mollusk Aplysia californica. To maximize RNA recoveries, optimized extraction solvents consisting of 10% methanol and chloroform were evaluated under dynamic and static extraction conditions. An MS-compatible RNA digestion buffer was developed to minimize the number of sample-transfer steps and facilitate the direct enzymatic digestion of extracted RNA within the sample collection tube. Compared to RNA extraction using a conventional phenol-chloroform method, the LMJ-based method provided a 3-fold greater coverage of the neuronal epitranscriptome for similar amounts of tissues and also produced mRNA of sufficient purity for reverse transcription polymerase chain reaction amplification. Using this approach, the expression of RNA-modifying enzymes in a given neuronal cell cluster can be characterized and simultaneously correlated with the LC-MS/MS analysis of RNA modifications within the same subset of neurons.
Figures


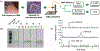
Similar articles
-
Characterizing RNA modifications in the central nervous system and single cells by RNA sequencing and liquid chromatography-tandem mass spectrometry techniques.Anal Bioanal Chem. 2023 Jul;415(18):3739-3748. doi: 10.1007/s00216-023-04604-y. Epub 2023 Feb 25. Anal Bioanal Chem. 2023. PMID: 36840809 Review.
-
Characterizing RNA Modifications in Single Neurons Using Mass Spectrometry.J Vis Exp. 2022 Apr 21;(182):10.3791/63940. doi: 10.3791/63940. J Vis Exp. 2022. PMID: 35532275 Free PMC article.
-
[Simultaneous determination of 30 antibiotics in soil by ultra-high performance liquid chromatography-tandem mass spectrometry].Se Pu. 2021 Aug;39(8):878-888. doi: 10.3724/SP.J.1123.2021.02019. Se Pu. 2021. PMID: 34212588 Free PMC article. Chinese.
-
Quantification of D-amino acids in the central nervous system of Aplysia californica by liquid chromatography/tandem mass spectrometry.Rapid Commun Mass Spectrom. 2007;21(1):73-7. doi: 10.1002/rcm.2803. Rapid Commun Mass Spectrom. 2007. PMID: 17133650
-
Androgen glucuronides analysis by liquid chromatography tandem-mass spectrometry: could it raise new perspectives in the diagnostic field of hormone-dependent malignancies?J Chromatogr B Analyt Technol Biomed Life Sci. 2013 Dec 1;940:24-34. doi: 10.1016/j.jchromb.2013.09.022. Epub 2013 Sep 27. J Chromatogr B Analyt Technol Biomed Life Sci. 2013. PMID: 24140653 Review.
Cited by
-
Rapid Determination of RNA Modifications in Consensus Motifs by Nuclease Protection with Ion-Tagged Oligonucleotide Probes and Matrix-Assisted Laser Desorption Ionization Mass Spectrometry.Genes (Basel). 2022 Jun 2;13(6):1008. doi: 10.3390/genes13061008. Genes (Basel). 2022. PMID: 35741770 Free PMC article.
-
Single-Neuron RNA Modification Analysis by Mass Spectrometry: Characterizing RNA Modification Patterns and Dynamics with Single-Cell Resolution.Anal Chem. 2021 Nov 2;93(43):14537-14544. doi: 10.1021/acs.analchem.1c03507. Epub 2021 Oct 21. Anal Chem. 2021. PMID: 34672536 Free PMC article.
-
Characterization of Neuronal RNA Modifications during Non-associative Learning in Aplysia Reveals Key Roles for tRNAs in Behavioral Sensitization.ACS Cent Sci. 2021 Jul 28;7(7):1183-1190. doi: 10.1021/acscentsci.1c00351. Epub 2021 Jul 18. ACS Cent Sci. 2021. PMID: 34345669 Free PMC article.
-
Characterizing RNA modifications in the central nervous system and single cells by RNA sequencing and liquid chromatography-tandem mass spectrometry techniques.Anal Bioanal Chem. 2023 Jul;415(18):3739-3748. doi: 10.1007/s00216-023-04604-y. Epub 2023 Feb 25. Anal Bioanal Chem. 2023. PMID: 36840809 Review.
-
Photocatalytic reduction-based liquid microjunction surface sampling-mass spectrometry for rapid in situ analysis of aromatic amines originating from azo dyes in packaging papers.Anal Bioanal Chem. 2021 Nov;413(26):6649-6660. doi: 10.1007/s00216-021-03631-x. Epub 2021 Sep 8. Anal Bioanal Chem. 2021. PMID: 34495385
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

