Current Status of SUMOylation Inhibitors
- PMID: 32778019
- PMCID: PMC8483067
- DOI: 10.2174/0929867327666200810135039
Current Status of SUMOylation Inhibitors
Abstract
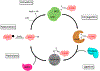
SUMOylation has emerged as an important post-translational modification that involves the covalent attachment of the Small Ubiquitin-like Modifier (SUMO) polypeptide to a lysine residue of a target protein. The enzymatic pathway of SUMOylation is very similar to ubiquitinylation and involves an activating enzyme, a conjugating enzyme, ligases, and deconjugating enzymes. SUMOylation modulates the function of a number of proteins associated with various pathways, and in fact, dysregulation of the SUMOylation pathway is observed in both cancer and neurological diseases. In many cancers, the SUMO enzymes are upregulated, and SUMO levels correlate directly with prognosis and disease progression. As a result, there has been an emphasis on the discovery and development of inhibitors of SUMOylation. In this review, the latest advances in SUMOylation inhibitors are described alongside the methods used to discover small molecule SUMOylation inhibitors, which include natural products, peptidomimetics, as well as synthetic derivatives identified via virtual screens.
Keywords: SUMO; cancer; enzyme inhibitors.; natural products; post-translational modifications; small-molecules; ubiquitin-like.
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Figures

Similar articles
-
Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells.J Biomed Sci. 2024 Jan 27;31(1):16. doi: 10.1186/s12929-024-01003-y. J Biomed Sci. 2024. PMID: 38280996 Free PMC article. Review.
-
Small-molecule inhibitors targeting small ubiquitin-like modifier pathway for the treatment of cancers and other diseases.Eur J Med Chem. 2022 Apr 5;233:114227. doi: 10.1016/j.ejmech.2022.114227. Epub 2022 Feb 28. Eur J Med Chem. 2022. PMID: 35247754 Review.
-
Molecular mechanisms in SUMO conjugation.Biochem Soc Trans. 2020 Feb 28;48(1):123-135. doi: 10.1042/BST20190357. Biochem Soc Trans. 2020. PMID: 31872228 Review.
-
E2-mediated small ubiquitin-like modifier (SUMO) modification of thymine DNA glycosylase is efficient but not selective for the enzyme-product complex.J Biol Chem. 2014 May 30;289(22):15810-9. doi: 10.1074/jbc.M114.572081. Epub 2014 Apr 21. J Biol Chem. 2014. PMID: 24753249 Free PMC article.
-
Mechanisms, regulation and consequences of protein SUMOylation.Biochem J. 2010 May 13;428(2):133-45. doi: 10.1042/BJ20100158. Biochem J. 2010. PMID: 20462400 Free PMC article. Review.
Cited by
-
Mechanisms and functions of SUMOylation in health and disease: a review focusing on immune cells.J Biomed Sci. 2024 Jan 27;31(1):16. doi: 10.1186/s12929-024-01003-y. J Biomed Sci. 2024. PMID: 38280996 Free PMC article. Review.
-
SUMOylation in Skeletal Development, Homeostasis, and Disease.Cells. 2022 Aug 31;11(17):2710. doi: 10.3390/cells11172710. Cells. 2022. PMID: 36078118 Free PMC article. Review.
-
Targeted inhibition of SUMOylation: treatment of tumors.Hum Cell. 2024 Sep;37(5):1347-1354. doi: 10.1007/s13577-024-01092-9. Epub 2024 Jun 10. Hum Cell. 2024. PMID: 38856883 Review.
-
Novel insights into the impact of the SUMOylation pathway in hematological malignancies (Review).Int J Oncol. 2021 Sep;59(3):73. doi: 10.3892/ijo.2021.5253. Epub 2021 Aug 9. Int J Oncol. 2021. PMID: 34368858 Free PMC article. Review.
-
SUMOylation and DeSUMOylation: Tug of War of Pain Signaling.Mol Neurobiol. 2024 Sep 14. doi: 10.1007/s12035-024-04478-w. Online ahead of print. Mol Neurobiol. 2024. PMID: 39276308 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

