Genomic Prediction of Agronomic Traits in Common Bean (Phaseolus vulgaris L.) Under Environmental Stress
- PMID: 32774338
- PMCID: PMC7381332
- DOI: 10.3389/fpls.2020.01001
Genomic Prediction of Agronomic Traits in Common Bean (Phaseolus vulgaris L.) Under Environmental Stress
Abstract
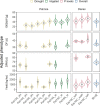
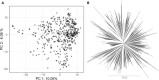
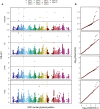
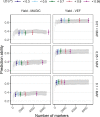
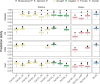
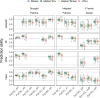
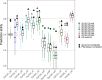
In plant and animal breeding, genomic prediction models are established to select new lines based on genomic data, without the need for laborious phenotyping. Prediction models can be trained on recent or historic phenotypic data and increasingly available genotypic data. This enables the adoption of genomic selection also in under-used legume crops such as common bean. Beans are an important staple food in the tropics and mainly grown by smallholders under limiting environmental conditions such as drought or low soil fertility. Therefore, genotype-by-environment interactions (G × E) are an important consideration when developing new bean varieties. However, G × E are often not considered in genomic prediction models nor are these models implemented in current bean breeding programs. Here we show the prediction abilities of four agronomic traits in common bean under various environmental stresses based on twelve field trials. The dataset includes 481 elite breeding lines characterized by 5,820 SNP markers. Prediction abilities over all twelve trials ranged between 0.6 and 0.8 for yield and days to maturity, respectively, predicting new lines into new seasons. In all four evaluated traits, the prediction abilities reached about 50-80% of the maximum accuracies given by phenotypic correlations and heritability. Predictions under drought and low phosphorus stress were up to 10 and 20% improved when G × E were included in the model, respectively. Our results demonstrate the potential of genomic selection to increase the genetic gain in common bean breeding. Prediction abilities improved when more phenotypic data was available and G × E could be accounted for. Furthermore, the developed models allowed us to predict genotypic performance under different environmental stresses. This will be a key factor in the development of common bean varieties adapted to future challenging conditions.
Keywords: common bean (Phaseolus vulgaris L.); drought; genome-wide association studies (GWAS); genomic selection; genotype × environment interactions; low phosphorus stress; plant breeding.
Copyright © 2020 Keller, Ariza-Suarez, de la Hoz, Aparicio, Portilla-Benavides, Buendia, Mayor, Studer and Raatz.
Figures
Similar articles
-
Genetic mapping for agronomic traits in a MAGIC population of common bean (Phaseolus vulgaris L.) under drought conditions.BMC Genomics. 2020 Nov 16;21(1):799. doi: 10.1186/s12864-020-07213-6. BMC Genomics. 2020. PMID: 33198642 Free PMC article.
-
Effective Use of Water and Increased Dry Matter Partitioned to Grain Contribute to Yield of Common Bean Improved for Drought Resistance.Front Plant Sci. 2016 May 12;7:660. doi: 10.3389/fpls.2016.00660. eCollection 2016. Front Plant Sci. 2016. PMID: 27242861 Free PMC article.
-
Novel SNP markers for flowering and seed quality traits in faba bean (Vicia faba L.): characterization and GWAS of a diversity panel.Front Plant Sci. 2024 Mar 6;15:1348014. doi: 10.3389/fpls.2024.1348014. eCollection 2024. Front Plant Sci. 2024. PMID: 38510437 Free PMC article.
-
Common bean proteomics: Present status and future strategies.J Proteomics. 2017 Oct 3;169:239-248. doi: 10.1016/j.jprot.2017.03.019. Epub 2017 Mar 25. J Proteomics. 2017. PMID: 28347863 Review.
-
Genomics, genetics and breeding of common bean in Africa: A review of tropical legume project.Plant Breed. 2019 Aug;138(4):401-414. doi: 10.1111/pbr.12573. Epub 2018 Apr 17. Plant Breed. 2019. PMID: 31728074 Free PMC article. Review.
Cited by
-
Genome-Wide Association Study and Genomic Prediction of Fusarium Wilt Resistance in Common Bean Core Collection.Int J Mol Sci. 2023 Oct 18;24(20):15300. doi: 10.3390/ijms242015300. Int J Mol Sci. 2023. PMID: 37894980 Free PMC article.
-
Genetic diversity and population structure of wild and cultivated Crotalaria species based on genotyping-by-sequencing.PLoS One. 2022 Sep 1;17(9):e0272955. doi: 10.1371/journal.pone.0272955. eCollection 2022. PLoS One. 2022. PMID: 36048841 Free PMC article.
-
Genetic Analyses and Genomic Predictions of Root Rot Resistance in Common Bean Across Trials and Populations.Front Plant Sci. 2021 Mar 12;12:629221. doi: 10.3389/fpls.2021.629221. eCollection 2021. Front Plant Sci. 2021. PMID: 33777068 Free PMC article.
-
riceExplorer: Uncovering the Hidden Potential of a National Genomic Resource Against a Global Database.Front Plant Sci. 2022 Apr 29;13:781153. doi: 10.3389/fpls.2022.781153. eCollection 2022. Front Plant Sci. 2022. PMID: 35574109 Free PMC article.
-
Genome-wide association and genomic prediction for iron and zinc concentration and iron bioavailability in a collection of yellow dry beans.Front Genet. 2024 Feb 6;15:1330361. doi: 10.3389/fgene.2024.1330361. eCollection 2024. Front Genet. 2024. PMID: 38380426 Free PMC article.
References
-
- Achard S. (2012). brainwaver: Basic wavelet analysis of multivariate time series with a visualisation and parametrisation using graph theory. R Project for Statistical Computing (R-Packages).
-
- Acosta-Pech R., Crossa J., de los Campos G., Teyssèdre S., Claustres B., Pérez-Elizalde S., et al. (2017). Genomic models with genotype × environment interaction for predicting hybrid performance: an application in maize hybrids. Theor. Appl. Genet. 130, 1431–1440. 10.1007/s00122-017-2898-0 - DOI - PubMed
-
- Assefa T., Assibi Mahama A., Brown A. V., Cannon E. K. S., Rubyogo J. C., Rao I. M., et al. (2019). A review of breeding objectives, genomic resources, and marker-assisted methods in common bean (Phaseolus vulgaris L.). Mol. Breed. 39, 20. 10.1007/s11032-018-0920-0 - DOI
LinkOut - more resources
Full Text Sources

