Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum
- PMID: 32764599
- PMCID: PMC7413343
- DOI: 10.1038/s41598-020-70061-7
Genome-wide identification and characterization of DCL, AGO and RDR gene families in Saccharum spontaneum
Abstract
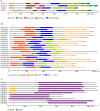
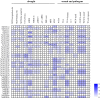
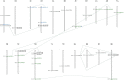
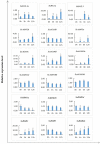
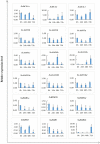
RNA silencing is a conserved mechanism in eukaryotic organisms to regulate gene expression. Argonaute (AGO), Dicer-like (DCL) and RNA-dependent RNA polymerase (RDR) proteins are critical components of RNA silencing, but how these gene families' functions in sugarcane were largely unknown. Most stress-resistance genes in modern sugarcane cultivars (Saccharum spp.) were originated from wild species of Saccharum, for example S. spontaneum. Here, we used genome-wide analysis and a phylogenetic approach to identify four DCL, 21 AGO and 11 RDR genes in the S. spontaneum genome (termed SsDCL, SsAGO and SsRDR, respectively). Several genes, particularly some of the SsAGOs, appeared to have undergone tandem or segmental duplications events. RNA-sequencing data revealed that four SsAGO genes (SsAGO18c, SsAGO18b, SsAGO10e and SsAGO6b) and three SsRDR genes (SsRDR2b, SsRDR2d and SsRDR3) tended to have preferential expression in stem tissue, while SsRDR5 was preferentially expressed in leaves. qRT-PCR analysis showed that SsAGO10c, SsDCL2 and SsRDR6b expressions were strongly upregulated, whereas that of SsAGO18b, SsRDR1a, SsRDR2b/2d and SsRDR5 was significantly depressed in S. spontaneum plants exposed to PEG-induced dehydration stress or infected with Xanthomonas albilineans, causal agent of leaf scald disease of sugarcane, suggesting that these genes play important roles in responses of S. spontaneum to biotic and abiotic stresses.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide identification of DCL, AGO and RDR gene families and their associated functional regulatory elements analyses in banana (Musa acuminata).PLoS One. 2021 Sep 2;16(9):e0256873. doi: 10.1371/journal.pone.0256873. eCollection 2021. PLoS One. 2021. PMID: 34473743 Free PMC article.
-
In silico identification and characterization of AGO, DCL and RDR gene families and their associated regulatory elements in sweet orange (Citrus sinensis L.).PLoS One. 2020 Dec 21;15(12):e0228233. doi: 10.1371/journal.pone.0228233. eCollection 2020. PLoS One. 2020. PMID: 33347517 Free PMC article.
-
Genome-wide identification and expression profiling of DREB genes in Saccharum spontaneum.BMC Genomics. 2021 Jun 17;22(1):456. doi: 10.1186/s12864-021-07799-5. BMC Genomics. 2021. PMID: 34139993 Free PMC article.
-
Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L.).BMC Genomics. 2020 Oct 2;21(1):685. doi: 10.1186/s12864-020-07076-x. BMC Genomics. 2020. PMID: 33008299 Free PMC article.
-
Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana.Cytogenet Genome Res. 2005;109(1-3):27-33. doi: 10.1159/000082378. Cytogenet Genome Res. 2005. PMID: 15753555 Review.
Cited by
-
Genome-Wide Identification and Characterization of Argonaute, Dicer-like and RNA-Dependent RNA Polymerase Gene Families and Their Expression Analyses in Fragaria spp.Genes (Basel). 2023 Jan 1;14(1):121. doi: 10.3390/genes14010121. Genes (Basel). 2023. PMID: 36672862 Free PMC article.
-
Genome-wide identification of DCL, AGO and RDR gene families and their associated functional regulatory elements analyses in banana (Musa acuminata).PLoS One. 2021 Sep 2;16(9):e0256873. doi: 10.1371/journal.pone.0256873. eCollection 2021. PLoS One. 2021. PMID: 34473743 Free PMC article.
-
Function and regulation of plant ARGONAUTE proteins in response to environmental challenges: a review.PeerJ. 2024 Mar 26;12:e17115. doi: 10.7717/peerj.17115. eCollection 2024. PeerJ. 2024. PMID: 38560454 Free PMC article. Review.
-
In silico identification and characterization of AGO, DCL and RDR gene families and their associated regulatory elements in sweet orange (Citrus sinensis L.).PLoS One. 2020 Dec 21;15(12):e0228233. doi: 10.1371/journal.pone.0228233. eCollection 2020. PLoS One. 2020. PMID: 33347517 Free PMC article.
-
Genome-wide identification of DCL, AGO and RDR gene families and their associated functional regulatory element analyses in sunflower (Helianthus annuus).PLoS One. 2023 Jun 9;18(6):e0286994. doi: 10.1371/journal.pone.0286994. eCollection 2023. PLoS One. 2023. PMID: 37294803 Free PMC article.
References
-
- Lopez-Gomollon S, Dalmay T. Recent patents in RNA silencing in plants: constructs, methods and applications in plant biotechnology. Recent Pat. DNA Gene Seq. 2010;4:155–166. - PubMed
-
- Sarkies P, Miska EA. Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat. Rev. Mol. Cell Biol. 2014;15:525–535. - PubMed
-
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. - PubMed
-
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 2013;64:137–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

