Selective Upregulation of CTLA-4 on CD8+ T Cells Restricted by HLA-B*35Px Renders them to an Exhausted Phenotype in HIV-1 infection
- PMID: 32760139
- PMCID: PMC7410205
- DOI: 10.1371/journal.ppat.1008696
Selective Upregulation of CTLA-4 on CD8+ T Cells Restricted by HLA-B*35Px Renders them to an Exhausted Phenotype in HIV-1 infection
Abstract
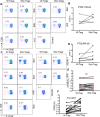
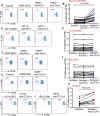
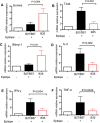
HLA-B*35Px is associated with HIV-1 disease rapid progression to AIDS. However, the mechanism(s) underlying this deleterious effect of this HLA allele on HIV-1 infection outcome has not fully understood. CD8+ T cells play a crucial role to control the viral replication but impaired CD8+ T cells represent a major hallmark of HIV-1 infection. Here, we examined the effector functions of CD8+ T cells restricted by HLA-B*35Px (HLA-B*35:03 and HLA-B*35:02), HLA-B*27/B57 and non-HLA-B*27/B57 (e.g. HLA-A*01, A*02, A*03, A*11, A*24, A*26, B*40, B*08, B*38, B*44). CD8+ T cells restricted by HLA-B*35Px exhibited an impaired phenotype compared with those restricted by HLA-B*27/B57 and even non-HLA-B*27/B57. CD8+ T cells restricted by non-HLA-B*27/B57 when encountered their cognate epitopes upregulated TIM-3 and thus became suppressed by regulatory T cells (Tregs) via TIM-3: Galectin-9 (Gal-9). Strikingly, CD8+ T cells restricted by HLA-B*35Px expressed fewer TIM-3 and therefore did not get suppressed by Tregs, which was similar to CD8+ T cells restricted by HLA-B*27/B57. Instead, CD8+ T cells restricted by HLA-B*35Px upon recognition of their cognate epitopes upregulated CTLA-4. The transcriptional and impaired phenotype (e.g. poor effector functions) of HIV-specific CD8+ T cells restricted by HLA-B*35 was related to persistent CTLA-4, elevated Eomes and blimp-1 but poor T-bet expression. As such, anti-CTLA-4 antibody, Ipilimumab, reversed the impaired proliferative capacity of antigen-specific CD8+ T cells restricted by HLA-B*35Px but not others. This study supports the concept that CD8+ T resistance to Tregs-mediated suppression is related to allele restriction rather than the epitope specificity. Our results aid to explain a novel mechanism for the inability of HIV-specific CD8+ T cells restricted by HLA-B*35Px to control viral replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Impact of a single HLA-A*24:02-associated escape mutation on the detrimental effect of HLA-B*35:01 in HIV-1 control.EBioMedicine. 2018 Oct;36:103-112. doi: 10.1016/j.ebiom.2018.09.022. Epub 2018 Sep 22. EBioMedicine. 2018. PMID: 30249546 Free PMC article.
-
Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection.AIDS. 2003 Dec 5;17(18):2581-91. doi: 10.1097/00002030-200312050-00005. AIDS. 2003. PMID: 14685052
-
The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences.J Virol. 2003 Jun;77(12):6889-98. doi: 10.1128/jvi.77.12.6889-6898.2003. J Virol. 2003. PMID: 12768008 Free PMC article.
-
Protective HLA-B57: T cell and natural killer cell recognition in HIV infection.Biochem Soc Trans. 2022 Oct 31;50(5):1329-1339. doi: 10.1042/BST20220244. Biochem Soc Trans. 2022. PMID: 36111814 Free PMC article. Review.
-
Molecular signatures of T-cell inhibition in HIV-1 infection.Retrovirology. 2013 Mar 20;10:31. doi: 10.1186/1742-4690-10-31. Retrovirology. 2013. PMID: 23514593 Free PMC article. Review.
Cited by
-
Galectin-9 expression defines exhausted T cells and impaired cytotoxic NK cells in patients with virus-associated solid tumors.J Immunother Cancer. 2020 Dec;8(2):e001849. doi: 10.1136/jitc-2020-001849. J Immunother Cancer. 2020. PMID: 33310773 Free PMC article.
-
Differential effects of age, sex and dexamethasone therapy on ACE2/TMPRSS2 expression and susceptibility to SARS-CoV-2 infection.Front Immunol. 2022 Nov 3;13:1021928. doi: 10.3389/fimmu.2022.1021928. eCollection 2022. Front Immunol. 2022. PMID: 36405732 Free PMC article.
-
CD71+ erythroid cells suppress T-cell effector functions and predict immunotherapy outcomes in patients with virus-associated solid tumors.J Immunother Cancer. 2023 May;11(5):e006595. doi: 10.1136/jitc-2022-006595. J Immunother Cancer. 2023. PMID: 37236637 Free PMC article.
-
Differential Signature of the Microbiome and Neutrophils in the Oral Cavity of HIV-Infected Individuals.Front Immunol. 2021 Nov 9;12:780910. doi: 10.3389/fimmu.2021.780910. eCollection 2021. Front Immunol. 2021. PMID: 34858437 Free PMC article.
-
Neutrophils promote T-cell activation through the regulated release of CD44-bound Galectin-9 from the cell surface during HIV infection.PLoS Biol. 2021 Aug 19;19(8):e3001387. doi: 10.1371/journal.pbio.3001387. eCollection 2021 Aug. PLoS Biol. 2021. PMID: 34411088 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

