Chemometric Models of Differential Amino Acids at the Navα and Navβ Interface of Mammalian Sodium Channel Isoforms
- PMID: 32756517
- PMCID: PMC7435598
- DOI: 10.3390/molecules25153551
Chemometric Models of Differential Amino Acids at the Navα and Navβ Interface of Mammalian Sodium Channel Isoforms
Abstract
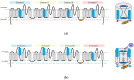
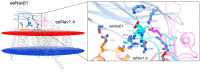
(1) Background: voltage-gated sodium channels (Navs) are integral membrane proteins that allow the sodium ion flux into the excitable cells and initiate the action potential. They comprise an α (Navα) subunit that forms the channel pore and are coupled to one or more auxiliary β (Navβ) subunits that modulate the gating to a variable extent. (2) Methods: after performing homology in silico modeling for all nine isoforms (Nav1.1α to Nav1.9α), the Navα and Navβ protein-protein interaction (PPI) was analyzed chemometrically based on the primary and secondary structures as well as topological or spatial mapping. (3) Results: our findings reveal a unique isoform-specific correspondence between certain segments of the extracellular loops of the Navα subunits. Precisely, loop S5 in domain I forms part of the PPI and assists Navβ1 or Navβ3 on all nine mammalian isoforms. The implied molecular movements resemble macroscopic springs, all of which explains published voltage sensor effects on sodium channel fast inactivation in gating. (4) Conclusions: currently, the specific functions exerted by the Navβ1 or Navβ3 subunits on the modulation of Navα gating remain unknown. Our work determined functional interaction in the extracellular domains on theoretical grounds and we propose a schematic model of the gating mechanism of fast channel sodium current inactivation by educated guessing.
Keywords: extracellular loops; homology modeling; hot spot prediction; interactome; protein-protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



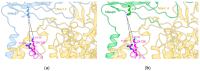
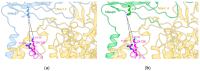



Similar articles
-
Molecular Pathology of Sodium Channel Beta-Subunit Variants.Front Pharmacol. 2021 Nov 19;12:761275. doi: 10.3389/fphar.2021.761275. eCollection 2021. Front Pharmacol. 2021. PMID: 34867379 Free PMC article.
-
Α- and β-subunit composition of voltage-gated sodium channels investigated with μ-conotoxins and the recently discovered μO§-conotoxin GVIIJ.J Neurophysiol. 2015 Apr 1;113(7):2289-301. doi: 10.1152/jn.01004.2014. Epub 2015 Jan 28. J Neurophysiol. 2015. PMID: 25632083 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
-
Backbone resonance assignments of complexes of apo human calmodulin bound to IQ motif peptides of voltage-dependent sodium channels NaV1.1, NaV1.4 and NaV1.7.Biomol NMR Assign. 2018 Oct;12(2):283-289. doi: 10.1007/s12104-018-9824-5. Epub 2018 May 4. Biomol NMR Assign. 2018. PMID: 29728980 Free PMC article.
-
Ubiquitylation of voltage-gated sodium channels.Handb Exp Pharmacol. 2014;221:231-50. doi: 10.1007/978-3-642-41588-3_11. Handb Exp Pharmacol. 2014. PMID: 24737239 Review.
Cited by
-
Molecular Pathology of Sodium Channel Beta-Subunit Variants.Front Pharmacol. 2021 Nov 19;12:761275. doi: 10.3389/fphar.2021.761275. eCollection 2021. Front Pharmacol. 2021. PMID: 34867379 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

