Mitochondrial morphology and activity regulate furrow ingression and contractile ring dynamics in Drosophila cellularization
- PMID: 32755438
- PMCID: PMC7851960
- DOI: 10.1091/mbc.E20-03-0177
Mitochondrial morphology and activity regulate furrow ingression and contractile ring dynamics in Drosophila cellularization
Abstract
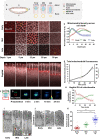
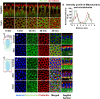
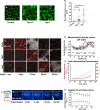
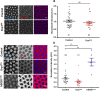
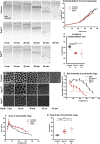
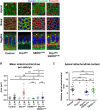
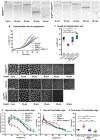
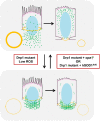
Mitochondria are maternally inherited in many organisms. Mitochondrial morphology and activity regulation is essential for cell survival, differentiation, and migration. An analysis of mitochondrial dynamics and function in morphogenetic events in early metazoan embryogenesis has not been carried out. In our study we find a crucial role of mitochondrial morphology regulation in cell formation in Drosophila embryogenesis. We find that mitochondria are small and fragmented and translocate apically on microtubules and distribute progressively along the cell length during cellularization. Embryos mutant for the mitochondrial fission protein, Drp1 (dynamin-related protein 1), die in embryogenesis and show an accumulation of clustered mitochondria on the basal side in cellularization. Additionally, Drp1 mutant embryos contain lower levels of reactive oxygen species (ROS). ROS depletion was previously shown to decrease myosin II activity. Drp1 loss also leads to myosin II depletion at the membrane furrow, thereby resulting in decreased cell height and larger contractile ring area in cellularization similar to that in myosin II mutants. The mitochondrial morphology and cellularization defects in Drp1 mutants are suppressed by reducing mitochondrial fusion and increasing cytoplasmic ROS in superoxide dismutase mutants. Our data show a key role for mitochondrial morphology and activity in supporting the morphogenetic events that drive cellularization in Drosophila embryos.
Figures
Similar articles
-
Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos.Mol Biol Cell. 2004 Feb;15(2):838-50. doi: 10.1091/mbc.e03-06-0440. Epub 2003 Dec 2. Mol Biol Cell. 2004. PMID: 14657248 Free PMC article.
-
Drp1-mediated mitochondrial fission regulates calcium and F-actin dynamics during wound healing.Biol Open. 2020 May 3;9(5):bio048629. doi: 10.1242/bio.048629. Biol Open. 2020. PMID: 32184231 Free PMC article.
-
A role for myosin II in mammalian mitochondrial fission.Curr Biol. 2014 Feb 17;24(4):409-14. doi: 10.1016/j.cub.2013.12.032. Epub 2014 Jan 30. Curr Biol. 2014. PMID: 24485837 Free PMC article.
-
The coordinated regulation of mitochondrial structure and function by Drp1 for mitochondrial quality surveillance.BMB Rep. 2019 Feb;52(2):109-110. doi: 10.5483/BMBRep.2019.52.2.032. BMB Rep. 2019. PMID: 30760382 Free PMC article. Review.
-
Apoptosis in Drosophila: which role for mitochondria?Apoptosis. 2016 Mar;21(3):239-51. doi: 10.1007/s10495-015-1209-y. Apoptosis. 2016. PMID: 26679112 Review.
Cited by
-
Labeling and Tracking Mitochondria with Photoactivation in Drosophila Embryos.Bio Protoc. 2022 Mar 5;12(5):e4347. doi: 10.21769/BioProtoc.4347. eCollection 2022 Mar 5. Bio Protoc. 2022. PMID: 35592607 Free PMC article.
-
Mitochondrial fusion regulates proliferation and differentiation in the type II neuroblast lineage in Drosophila.PLoS Genet. 2022 Feb 14;18(2):e1010055. doi: 10.1371/journal.pgen.1010055. eCollection 2022 Feb. PLoS Genet. 2022. PMID: 35157701 Free PMC article.
-
Mitochondrial morphology dynamics and ROS regulate apical polarity and differentiation in Drosophila follicle cells.Development. 2024 Mar 1;151(5):dev201732. doi: 10.1242/dev.201732. Epub 2024 Feb 29. Development. 2024. PMID: 38345270 Free PMC article.
-
Membrane-actin interactions in morphogenesis: Lessons learned from Drosophila cellularization.Semin Cell Dev Biol. 2023 Jan 15;133:107-122. doi: 10.1016/j.semcdb.2022.03.028. Epub 2022 Apr 5. Semin Cell Dev Biol. 2023. PMID: 35396167 Free PMC article. Review.
-
Using Expansion Microscopy to Physically Enlarge Whole-Mount Drosophila Embryos for Super-Resolution Imaging.J Vis Exp. 2023 Apr 28;(194):10.3791/64662. doi: 10.3791/64662. J Vis Exp. 2023. PMID: 37184263 Free PMC article.
References
-
- Afshar K, Stuart B, Wasserman SA (2000). Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development , 1887–1897. - PubMed
-
- Bavister BD, Squirrell JM (2000). Mitochondrial distribution and function in oocytes and early embryos. Hum Reprod (Suppl 2), 189–198. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

