Structure-guided screening strategy combining surface plasmon resonance with nuclear magnetic resonance for identification of small-molecule Argonaute 2 inhibitors
- PMID: 32735606
- PMCID: PMC7394379
- DOI: 10.1371/journal.pone.0236710
Structure-guided screening strategy combining surface plasmon resonance with nuclear magnetic resonance for identification of small-molecule Argonaute 2 inhibitors
Abstract
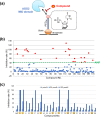
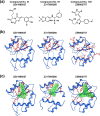
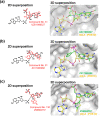
Argonaute (AGO) proteins are the key component of the RNA interference machinery that suppresses gene expression by forming an RNA-induced silencing complex (RISC) with microRNAs (miRNAs). Each miRNA is involved in various cellular processes, such as development, differentiation, tumorigenesis, and viral infection. Thus, molecules that regulate miRNA function are expected to have therapeutic potential. In addition, the biogenesis of miRNA is a multistep process involving various proteins, although the complete pathway remains to be elucidated. Therefore, identification of molecules that can specifically modulate each step will help understand the mechanism of gene suppression. To date, several AGO2 inhibitors have been identified. However, these molecules were identified through a single screening method, and no studies have specifically evaluated a combinatorial strategy. Here, we demonstrated a combinatorial screening (SCR) approach comprising an in silico molecular docking study, surface plasmon resonance (SPR) analysis, and nuclear magnetic resonance (NMR) analysis, focusing on the strong binding between the 5'-terminal phosphate of RNA and the AGO2 middle (MID) domain. By combining SPR and NMR, we identified binding modes of amino acid residues binding to AGO2. First, using a large chemical library (over 6,000,000 compounds), 171 compounds with acidic functional groups were screened using in silico SCR. Next, we constructed an SPR inhibition system that could analyze only the 5'-terminal binding site of RNA, and nine molecules that strongly bound to the AGO2 MID domain were selected. Finally, using NMR, three molecules that bound to the desired site were identified. The RISC inhibitory ability of the "hit" compounds was analyzed in human cell lysate, and all three hit compounds strongly inhibited the binding between double-stranded RNA and AGO2.
Conflict of interest statement
TH, AS, YT, HM, JS, and FS are affiliated with the commercial company, Kyowa Kirin. There are no patents, products in development, or marketed products to declare. This does not alter the authors’ adherence to all PLOS ONE policies on sharing data and materials.
Figures
Similar articles
-
Surface Plasmon Resonance: A Useful Strategy for the Identification of Small Molecule Argonaute 2 Protein Binders.Methods Mol Biol. 2017;1517:223-237. doi: 10.1007/978-1-4939-6563-2_16. Methods Mol Biol. 2017. PMID: 27924486
-
Small molecule inhibition of RISC loading.ACS Chem Biol. 2012 Feb 17;7(2):403-10. doi: 10.1021/cb200253h. Epub 2011 Nov 11. ACS Chem Biol. 2012. PMID: 22026461 Free PMC article.
-
A combination of 19F NMR and surface plasmon resonance for site-specific hit selection and validation of fragment molecules that bind to the ATP-binding site of a kinase.Bioorg Med Chem. 2018 May 1;26(8):1929-1938. doi: 10.1016/j.bmc.2018.02.041. Epub 2018 Feb 22. Bioorg Med Chem. 2018. PMID: 29510947
-
SPR-based fragment screening: advantages and applications.Curr Top Med Chem. 2007;7(16):1630-42. doi: 10.2174/156802607782341073. Curr Top Med Chem. 2007. PMID: 17979772 Review.
-
Intracellular localization and routing of miRNA and RNAi pathway components.Curr Top Med Chem. 2012;12(2):79-88. doi: 10.2174/156802612798919132. Curr Top Med Chem. 2012. PMID: 22196276 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

