Genetic Variation of Multiple Serotypes of Enteroviruses Associated with Hand, Foot and Mouth Disease in Southern China
- PMID: 32725479
- PMCID: PMC7385209
- DOI: 10.1007/s12250-020-00266-7
Genetic Variation of Multiple Serotypes of Enteroviruses Associated with Hand, Foot and Mouth Disease in Southern China
Abstract
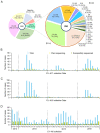
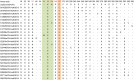
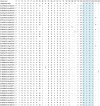
Enteroviruses (EVs) species A are a major public health issue in the Asia-Pacific region and cause frequent epidemics of hand, foot and mouth disease (HFMD) in China. Mild infections are common in children; however, HFMD can also cause severe illness that affects the central nervous system. To molecularly characterize EVs, a prospective HFMD virological surveillance program was performed in China between 2013 and 2016. Throat swabs, rectal swabs and stool samples were collected from suspected HFMD patients at participating hospitals. EVs were detected using generic real-time and nested reverse transcription-polymerase chain reactions (RT-PCRs). Then, the complete VP1 regions of enterovirus A71 (EV-A71), coxsackievirus A16 (CVA16) and CVA6 were sequenced to analyze amino acid changes and construct a viral molecular phylogeny. Of the 2836 enrolled HFMD patients, 2,517 (89%) were EV positive. The most frequently detected EVs were CVA16 (32.5%, 819), CVA6 (31.2%, 785), and EV-A71 (20.4%, 514). The subgenogroups CVA16_B1b, CVA6_D3a and EV-A71_C4a were predominant in China and recombination was not observed in the VP1 region. Sequence analysis revealed amino acid variations at the 30, 29 and 44 positions in the VP1 region of EV-A71, CVA16 and CVA6 (compared to the respective prototype strains BrCr, G10 and Gdula), respectively. Furthermore, in 21 of 24 (87.5%) identified EV-A71 samples, a known amino acid substitution (D31N) that may enhance neurovirulence was detected. Our study provides insights about the genetic characteristics of common HFMD-associated EVs. However, the emergence and virulence of the described mutations require further investigation.
Keywords: Coxsackievirus A16 (CVA16); Coxsackievirus A6 (CVA6); Enterovirus A71 (EV-A71); Enteroviruses (EVs); Hand, foot and mouth disease (HFMD).
Conflict of interest statement
HJY has received investigator-initiated research funding from Sanofi Pasteur, GlaxoSmithKline, and Yichang HEC Changjiang Pharmaceutical Company, none of which is related to HFMD and enteroviruses.
Figures




Similar articles
-
Genetic characteristics of Coxsackievirus A6 from children with hand, foot and mouth disease in Beijing, China, 2017-2019.Infect Genet Evol. 2022 Dec;106:105378. doi: 10.1016/j.meegid.2022.105378. Epub 2022 Oct 17. Infect Genet Evol. 2022. PMID: 36257478
-
Clinical features of hand, foot and mouth disease caused by Coxsackievirus A6 in Xi'an, China, 2013-2019: A multicenter observational study.Acta Trop. 2024 Sep;257:107310. doi: 10.1016/j.actatropica.2024.107310. Epub 2024 Jun 30. Acta Trop. 2024. PMID: 38955319
-
Genetic characterization of enterovirus strains identified in Hand, Foot and Mouth Disease (HFMD): Emergence of B1c, C1 subgenotypes, E2 sublineage of CVA16, EV71 and CVA6 strains in India.Infect Genet Evol. 2017 Oct;54:192-199. doi: 10.1016/j.meegid.2017.05.024. Epub 2017 Jun 1. Infect Genet Evol. 2017. PMID: 28577914
-
Atypical hand-foot-mouth disease in Belém, Amazon region, northern Brazil, with detection of coxsackievirus A6.J Clin Virol. 2020 May;126:104307. doi: 10.1016/j.jcv.2020.104307. Epub 2020 Mar 6. J Clin Virol. 2020. PMID: 32302950 Review.
-
Seroprevalence of coxsackievirus A6 and enterovirus A71 infection in humans: a systematic review and meta-analysis.Arch Virol. 2023 Jan 7;168(2):37. doi: 10.1007/s00705-022-05642-0. Arch Virol. 2023. PMID: 36609748 Free PMC article. Review.
Cited by
-
Hand, Foot, and Mouth Disease: A Narrative Review.Recent Adv Inflamm Allergy Drug Discov. 2022;16(2):77-95. doi: 10.2174/1570180820666221024095837. Recent Adv Inflamm Allergy Drug Discov. 2022. PMID: 36284392 Review.
-
Diagnostic performance of different specimens in detecting enterovirus A71 in children with hand, foot and mouth disease.Virol Sin. 2023 Apr;38(2):268-275. doi: 10.1016/j.virs.2022.11.004. Epub 2022 Nov 10. Virol Sin. 2023. PMID: 36371008 Free PMC article.
-
Seroprevalence of neutralizing antibodies against HFMD associated enteroviruses among healthy individuals in Shanghai, China, 2022.Virol Sin. 2024 Aug;39(4):694-698. doi: 10.1016/j.virs.2024.05.008. Epub 2024 May 25. Virol Sin. 2024. PMID: 38801978 Free PMC article.
-
Molecular Evolutionary Dynamics of Coxsackievirus A6 Causing Hand, Foot, and Mouth Disease From 2021 to 2023 in China: Genomic Epidemiology Study.JMIR Public Health Surveill. 2024 Jul 31;10:e59604. doi: 10.2196/59604. JMIR Public Health Surveill. 2024. PMID: 39087568 Free PMC article.
-
Identification of specific and shared epitopes at the extreme N-terminal VP1 of Coxsackievirus A4, A2 and A5 by monoclonal antibodies.Virus Res. 2023 Apr 15;328:199074. doi: 10.1016/j.virusres.2023.199074. Epub 2023 Mar 1. Virus Res. 2023. PMID: 36805409 Free PMC article.
References
-
- Anh NT, Nhu LNT, Van HMT, Hong NTT, Thanh TT, Hang VTT, Ny NTH, Nguyet LA, Phuong TTL, Nhan LNT, Hung NT, Khanh TH, Tuan HM, Viet HL, Nam NT, Viet DC, Qui PT, Wills B, Sabanathan S, Chau NVV, Thwaites L, Rogier van Doorn H, Thwaites G, Rabaa MA, Van Tan L. Emerging coxsackievirus a6 causing hand, foot and mouth disease, vietnam. Emerg Infect Dis. 2018;24:654–662. - PMC - PubMed
-
- Bian L, Wang Y, Yao X, Mao Q, Xu M, Liang Z. Coxsackievirus A6: a new emerging pathogen causing hand, foot and mouth disease outbreaks worldwide. Expert Rev Anti Infect Ther. 2015;13:1061–1071. - PubMed
-
- Chan YF, Wee KL, Chiam CW, Khor CS, Chan SY, Amalina WM, Sam IC. Comparative genetic analysis of VP4, VP1 and 3D gene regions of enterovirus 71 and coxsackievirus A16 circulating in malaysia between 1997–2008. Trop Biomed. 2012;29:451–466. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

